Abstract
Assessment of structural and functional changes of mitochondria is vital for biomedical research as mitochondria are the power plants essential for biological processes and tissue/organ functions. Others and we have developed a novel reporter gene, pMitoTimer, which codes for a redox sensitive mitochondrial targeted protein that switches from green fluorescence protein (GFP) to red fluorescent protein (DsRed) when oxidized. It has been shown in transfected cells, transgenic C. elegans and Drosophila m., as well as somatically transfected adult skeletal muscle that this reporter gene allows quantifiable assessment of mitochondrial structure, oxidative stress, and lysosomal targeting of mitochondria-containing autophagosomes. Here, we generated CAG-CAT-MitoTimer transgenic mice using a transgene containing MitoTimer downstream of LoxP-flanked bacterial chloramphenicol acetyltransferase (CAT) gene with stop codon under the control of the cytomegalovirus (CMV) enhancer fused to the chicken β-actin promoter (CAG). When CAG-CAT-MitoTimer mice were crossbred with various tissue-specific (muscle, adipose tissue, kidney, and pancreatic tumor) or global Cre transgenic mice, the double transgenic offspring showed MitoTimer expression in tissue-specific or global manner. Lastly, we show that hindlimb ischemia-reperfusion caused early, transient increases of mitochondrial oxidative stress, mitochondrial fragmentation and lysosomal targeting of autophagosomes containing mitochondria as well as a later reduction of mitochondrial content in skeletal muscle along with mitochondrial oxidative stress in sciatic nerve. Thus, we have generated conditional MitoTimer mice and provided proof of principle evidence of their utility to simultaneously assess mitochondrial structure, oxidative stress, and mitophagy in vivo in a tissue-specific, controllable fashion.
1. Introduction
Mitochondrial biogenesis, dynamics (reorganization of the mitochondrial reticulum through fission and fusion), and mitophagy (degradation of damaged/dysfunctional regions via the lysosomal degradation system), collectively maintain mitochondrial quality in response to stressors (Drake et al., 2015; Kroemer et al., 2010). Impaired mitochondrial quality is a hallmark of many chronic diseases, including, but not limited to, cardiovascular disease, diabetes, neurological disorders and aging (Knuppertz and Osiewacz, 2016; Matic et al., 2015; Wanagat and Hevener, 2016; Vásquez-Trincado et al., 2016). The importance of mitochondrial quality maintenance in disease susceptibility and progression across multiple different tissue/organ systems warrants a comprehensive, in-depth understanding of how mitochondrial quality is regulated in response to stressors in vivo.
One way to determine mitochondrial stress is to measure production and/or consequence of excessive reactive oxygen species (ROS). Multiple fluorescent-based reporter genes have been developed to assess mitochondria ROS (mtROS) emission. Mitochondria-targeted roGFP, HyPer, and mt-cpYFP detect redox state and H2O2 in the mitochondria (Roma et al., 2012; Wang et al., 2008; Wolf et al., 2008) nearly real-time; however, these reporter genes are generally not suitable for assessing the accompanied processes (i.e. biogenesis, fission/fusion, and mitophagy) that are often activated under various conditions of stress. Recently, Sun et al. developed a transgenic mouse expressing a mitochondria-targeted, pH-sensitive fluorophore, mt-Kiema, that changes fluorescence from green to red when engulfed in lysosomes allowing assessment of mitophagy (Sun et al., 2015). Transfection of mitochondria-targeted GFP has been used to assess mitochondrial dynamics and morphology (Toyama et al., 2016), but its in vivo utility is unclear (Hodneland Nilsson et al., 2015; Roma et al., 2012). The mito-Dendra2 mouse, which expresses a photo-activatable mitochondrial transgene (PhAM), has been shown to circumvent the technical challenge of tracking mitochondrial dynamics in vivo (Pham et al., 2012). Finally, recent development of a mouse model that combined these two principles opened the possibility of simultaneous assessment of mitophagy and mitochondrial dynamics in vivo (mito-QC) (Mcwilliams et al., 2016). An animal model for simultaneous assessment of mitochondrial stress, content, dynamics, and mitophagy in various tissues/organs in vivo would significantly improve our capability to dissect the molecular mechanisms of various physiological and pathological processes.
Others and we have developed novel pMitoTimer reporter genes (Call et al., 2017; Laker et al., 2017; Laker et al., 2014; Verkhusha et al., 2004; Yarbrough et al., 2001) from a fluorescent reporter gene, pTimer, which encodes DsRed1-E5 that fluoresces as GFP when newly synthesized and irreversibly shifts to DsRed upon oxidation at Tyr-67 (Terskikh, 2000). MitoTimer protein targets to mitochondria due to the addition of mitochondrial targeting sequence to the N-terminus of the MitoTimer coding region. We have demonstrated in transfected cells, transgenic C. elegans, and Drosophila m., as well as somatically transfected adult mouse skeletal muscle that MitoTimer allows quantifiable assessment of mitochondrial structure, oxidative stress, and lysosomal targeting of autophagosomes containing mtiochondria (Call et al., 2017; Laker et al., 2017; Laker et al., 2014). To fully take advantage of this reporter gene technology, we endeavored to generate and characterize a novel conditional MitoTimer reporter mouse line and test its utility for simultaneous assessment of mitochondrial structure, oxidative stress, and mitophagy in different tissues/organs in vivo.
2. Materials & methods
2.1. Plasmid DNA construct
We constructed pCAG-CAT-MitoTimer by inserting 700 bp BamHI-NotI fragments of pMitoTimer into the 6.4-kb plasmid DNA (digested with BamH1-NotI) containing the CMV enhanced β-actin promoter (a generous gift from Drs. Eric Olson and Beverly Rothermel at UT Southwestern).
2.2. Cell transfection
Immortalized C2C12 mouse myoblasts were co-transfected with pCAG-CAT-MitoTimer and CMV-Cre (Addgene) or an empty vector, pCI-Neo, using lipofectamine 3000 (ThermoFisher L3000015) following the manufacturer's protocol.
2.3. Animals
All animal procedures were conducted under the approval of the Institutional Animal Care and Use Committee of the University of Virginia. Isolated DNA fragment digestion of pCAG-CAT-MitoTimer, containing the CAG-CAT-MitoTimer expression unit (7.1 kb), was used for microinjection of male pronuclei of fertilized oocytes (C57BL/6 J) at the Genetically Engineered Murine Model Core of University of Virginia. One F0 was identified and used to breed with wild type C57BL/6 J mice to generate a transgenic line. F0 and F1 progenies were genotyped by PCR using designed primers (forward: 5′-GAGTTCATGCGCTTCAAGGT-3′, reverse: 3′-GAGGTGATGTCCAGCTTGGT-5′). To induce MitoTimer expression in tissue-specific or global manner, CAG-CAT-MitoTimer mice were crossbred with different transgenic lines with tissue-specific or globally active promoters driving Cre expression. To generate muscle-specific MitoTimer mice, we crossbred MCK-Cre (Jackson Laboratory) with CAG-CAT-MitoTimer mice (MCK-Cre;CAG-CAT-MitoTimer). To generate kidney proximal tubule-specific MitoTimer mice, we crossbred PepCK-Cre (Higgins et al., 2007) with CAG-CAT-MitoTimer mice (PepCK-Cre;CAG-CAT-MitoTimer). To create adipose tissue-specific inducible MitoTimer mice, we crossbred adiponectin-rtTA-TRE-Cre (Wang et al., 2015) with CAG-CAT-MitoTimer mice (adiponectin-rtTA-TRE-Cre;CAG-CAT-MitoTimer) and fed them with doxycycline-chow (200 mg/kg) for 2 weeks to induce MitoTimer expression in the adipose tissues. Pancreatic ductal adenocarcinoma MitoTimer mice were generated by crossing LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-CreERTg/+ (Friedlander et al., 2009) with CAG-CAT-MitoTimer mice (LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-CreERTg/+;CAG-CAT-MitoTimer). MitoTimer expression in pancreatic ductal adenocarcinoma was induced by i.p. injection of tamoxifen (9 mg/40 g body weight at postnatal days 22, 24 and 26). Global inducible MitoTimer mice were generated by crossbreeding between CAG-CreERT2 (Jackson Laboratory) with CAG-CAT-MitoTimer mice (CAG-CreERT2;CAG-CAT-MitoTimer). MitoTimer expression was induced by 7 days of tamoxifen injection (40 mg/kg, i.p.). Mice were used in experiments at least 3 days following the last injection of tamoxifen.
2.4. Pancreatic tumor cell isolation
Tumor cells were isolated as previously described with minor modifications (Ju et al., 2015). Briefly, at necropsy, tumor tissue was finely minced and incubated with Collagenase Type IV (ThermoFisher 17,104,019) at 2 mg/ml in DMEM (Invitrogen 11,965,092) for 30 min at 37 °C. The cell suspension and remaining small tumor fragments were rinsed and then resuspended in DMEM containing 10% FBS (Seradigm 1500) before being plated in 10 cm dishes. The cells were incubated at 5%CO2 at 37 °C.
2.5. Hindlimb ischemia-reperfusion injury
Acute ischemia-reperfusion (IR) injury was induced as previously described (Call et al., 2017), and identical between different mouse models. Briefly, under anesthesia an 1/8 in. 4-oz orthodontic rubber-band (DENTSPLY GAC International Inc. 11-102-03) was placed around the femur ~ 20 cm above the knee using McGivney Hemorroidal Ligator. After 1 h, the rubberband was removed to initiate reperfusion.
2.6. Immunoblotting
Whole muscle homogenates were obtained from the gastrocnemius muscle (GA), heart (HT) and liver (LV), resolved by electrophoresis on a SDS-page gel (30 μg/lane), then transferred to a nitrocellulose membrane (Odyssey 956-31092). After blocking in 5% milk in Tris-buffered saline with Tween 20 (TBST), the membrane was incubated overnight with rabbit anti-dsRED (Clontech 632496) diluted 1:1000 followed by incubation with goat anti-rabbit 680 (ThermoFisher SA5-35571) for 1 h with TBST washes in between and after. The membrane was scanned using Licor (Odyssey).
2.7. Confocal imaging
Tissues were fixed and prepared for either whole mounting or cryosectioning for MitoTimer confocal microscopy using FLUOVIEW Ver.4.2a software (Olympus) as previously described (Laker et al., 2014). MitoTimer was detected by the green (ex/em 488/518 nm) and red (ex/em 543/572 nm) channels. Identical acquisition parameters were used for every sample of the same tissue type.
2.8. MitoTimer analysis
MitoTimer Red:Green ratio and mitochondrial content was analyzed using a custom Matlab-based algorithm as previously described (Laker et al., 2014). Percent fiber with mitochondrial fragmentation was analyzed manually in an identity-blinded fashion.
2.9. Statistical analysis
All results are presented as means ± SE. Two-tailed unpaired t-test was used to compare MitoTimer Red:Green ratio, percentage mitochondria per fiber area, number of pure red puncta and percentage of fibers with fragmented mitochondria between sham treated and IR groups. For the time course studies, one-way ANOVA was employed followed by Tukey's multiple comparisons post hoc test to locate the difference once a statistically significant interaction was reached. P < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Development of pCAG-CAT-MitoTimer DNA construct and generation of transgenic mice
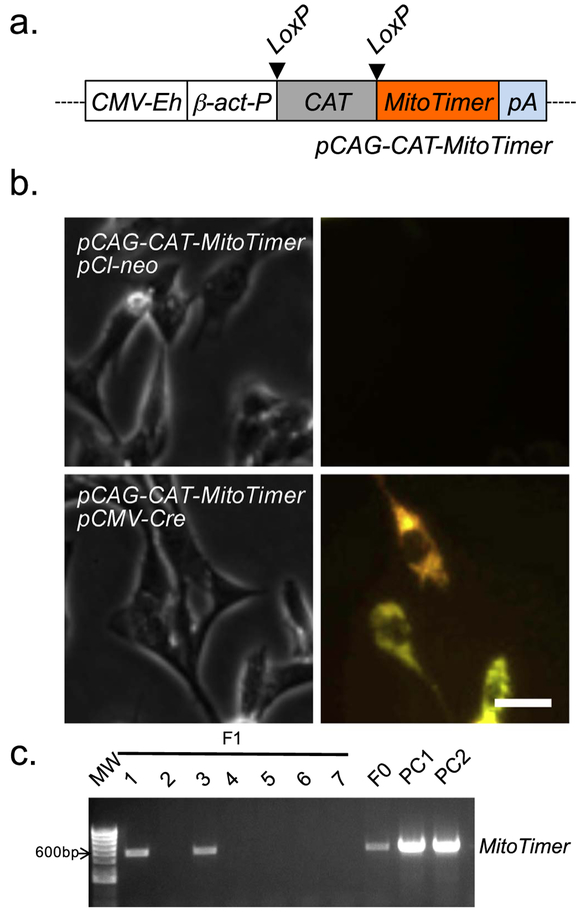
In order to generate conditional MitoTimer transgenic mice, we subcloned the MitoTimer coding region into an expression vector downstream from LoxP-flanked chloramphenicol acetyltransferase (CAT) coding sequence with a stop codon under the control of cytomegalovirus (CMV) enhanced β-actin promoter (Wang et al., 2015) that we named pCAG-CAT-MitoTimer (Fig. 1a). To ensure that this expression construct was correctly generated with inducible expression, we checked the structure of the plasmid DNA by combinatory restriction enzyme digestions (not shown) and co-transfected C2C12 myoblasts with pCAG-CAT-MitoTimer and pCMV-Cre (constitutive expression of Cre recombinase) using empty vector pCI-neo as the negative control. We confirmed MitoTimer expression via confocal microscopy only in cells that were co-transfected with pCMV-Cre, but not with empty vector (Fig. 1b). These findings confirmed the inducibility of the pCAG-CAT-MitoTimer expression vector in transiently transfected cells in culture dependent on the expression of Cre recombinase. We then proceeded to generate MitoTimer transgenic mice by microinjection of male pronuclei of fertilized oocytes (in C57BL/6 J background) with isolated DNA fragment containing the CAG-CAT-MitoTimer expression unit (Fig. 1a). Multiple F1 progenies were obtained by crossbreeding the F0 CAG-CAT-MitoTimer mouse with wild type C57BL/6 mice, confirming germline transmission of the transgene to offspring.
Fig. 1.
Development of pCAG-CAT-MitoTimer construct. A plasmid DNA containing conditional expression unit of MitoTimer was constructed by genetic engineering and used for conditional expression In cultured cells to confirm its inducibility. (a) A schematic presentation of the recombinant pCAG-CAT-Mitotimer construct generated by subcloning MitoTimer coding region Into a plasmid DNA containing the CMV enhanced β-actin promoter; (b) Representative confocal Images of C2C12 cells co-transfected of pCAG-CAT-MitoTimer with either empty vector pCI-neo or pCMV-Cre. MitoTimer expression was only detected In the presence of Cre; and (c) Genotyping of the F1 progeny following the crossbredding of CAG-CAT-MitoTimer mice with wild type (WT) mice. pCAG-CAT-MitoTimer and isolated DNA fragment of CAG-CAT-MitoTimer expression unit were Included to serve as positive controls 1 and 2 (PC1 and PC2), respectively.
3.2. Conditional expression of MitoTimer following crossbreeding of CAG-CAT-MitoTimer with various Cre transgenic lines
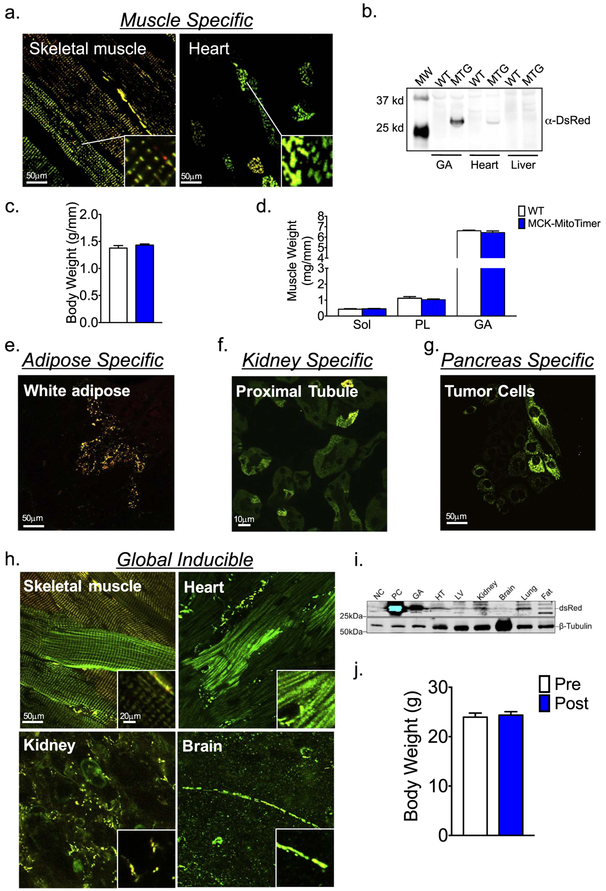
We obtained muscle-specific MitoTimer mice by crossbreeding CAG-CAT-MitoTimer mice with transgenic Cre mice driven by the muscle creatine kinase (MCK) promoter. We confirmed via both western blot and confocal microscopy that these mice expressed MitoTimer in all skeletal muscle fibers with some expression in cardiac myocytes (Fig. 2a & b). These findings are consistent with previous findings that the MCK promoter is specific for adult skeletal muscle, but with leaky activity in adult cardiac myocytes (Bruning et al., 1998). Importantly, expression of MCK-MitoTimer did not affect body weight or muscle wet weight (Fig. 2c & d), suggesting that mitochondrial metabolism was not impacted by MitoTimer.
Fig. 2.
Generation of MitoTimer transgenic mice. A transgenic mouse line was generated by pronclear injection of isolated DNA fragment containing the conditional MitoTimer expression unit as illustrated in Fig. 1a. (a) Representative confocal images of MitoTimer expression in the skeletal muscle and heart from muscle specific MitoTimer mice (MTG; MCK-Cre;CAG-CAT-MitoTimer); (b) Immunoblot analysis of MitoTimer protein expression gastrocnemius muscle (GA), heart and liver tissues in MTG and the wild type littermates (WT); (c) Body weight was not different between MCK-MitoTimer and their WT littermates; (d) Muscle weights for Soleus (Sol, Plantaris (PL)), and Gastrocnemius (GA), normalized to tibia length, was not different between MCK-MitoTimer and their WT littermates; (e) Representative confocal images of MitoTimer expression in white adipose tissue of adipose tissue-specific inducible MitoTimer mice (adiponectin-rtTA-TRE-Cre;CAG-CAT-MitoTimer); (f) Representative confocal images of MitoTimer expression in kidney proximal tubule-specific mice (PepCK-Cre;CAG-CAT-MitoTimer); (g) Representative confocal images of MitoTimer expression in primary cells isolated from pancreatic tumor cells from pancreatic ductal adenocarcinoma MitoTimer mice (LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-CreERTg/+;CAG-CAT-MitoTimer); (h) Representative confocal images of MitoTimer expression in skeletal muscle, heart, kidney, and brain in global inducible MitoTimer mice (CAG-CreERT2;CAG-CAT-MitoTimer); and (i) Immunoblot analysis of different tissues. Negative (NC) and positive (PC) controls are homogenates of the GA muscle from wild type mice and MTG mice, respectively. (j) Body weights in MTG mice was not affected by 7 days of tamoxifen injection.
When CAG-CAT-MitoTimer mice were crossbred adiponectin-rtTA-TRE-Cre mice, the double transgenic offspring showed MitoTimer expression revealing mitochondrial network in white adipose tissue following 2 weeks of feeding with doxycline-chow (Fig. 2e). Similarly, when crossbred with PepCK-Cre mice, the double transgenic offspring showed MitoTimer expression in the kidney proximal tubules (Fig. 2f). Finally, when crossed into LSL-KrasG12D/+;Trp53flox/flox;Pdx-1-CreERTg/+ background, MitoTimer expression was detected in isolated pancreatic ductal tumor cells (Fig. 2g). These findings provide convincing evidence that CAG-CAT-MitoTimer mice can be used to generate conditional MitoTimer reporter mice in different tissues/organs even with inducible expressions, thus allowing precise control of the expression in any tissue at any time across the lifespan.
We then obtained global, inducible MitoTimer mice by crossing CAG-CAT-MitoTimer mice with transgenic mice harboring the tamoxifen-inducible CAG-CreERT2 gene. Following 7 days of daily injections of tamoxifen, we observed MitoTimer expression in the skeletal muscle, heart, brain, kidney, liver, lung, and fat (Fig. 2h and i), which was not present in any tissue prior to tamoxifen treatment (data not shown). Expression of MitoTimer across tissues did not affect body weight (Fig. 2j), further supporting that MitoTimer does not impact metabolism. Importantly, the varying mitochondrial reticulum architecture between multiple tissue-specific MitoTimer mice (Fig. 2) is consistent with previous observations (Fu et al., 2017; Mcwilliams et al., 2016). We have previously demonstrated that MitoTimer protein expression under the control of a constitutively active promoter is suitable for detection of cumulative changes of mitochondrial structure and oxidative stress (Laker et al., 2014), which posits an advantage over other transient methodologies (Roma et al., 2012; Wang et al., 2008; Wolf et al., 2008). Although we have not fully characterized the identities of cells in various tissues/organs of the global MitoTimer reporter mice, the fact that skeletal muscle and cardiac myocytes showed typical mitochondrial network structure suggests that the confocal microscopy images of various tissues represent the authentic mitochondrial network structures, but we acknowledge that the respective system in question needs to be closely considered when interpreting results due to possible tissue-specific responses in some mitochondrial processes (Glancy et al., 2017). Thus, these transgenic models have a great potential for integrative assessments of mitochondrial structure and health under physiological and pathological conditions.
3.3. MitoTimer transgenic mice are sensitive to pathological stress
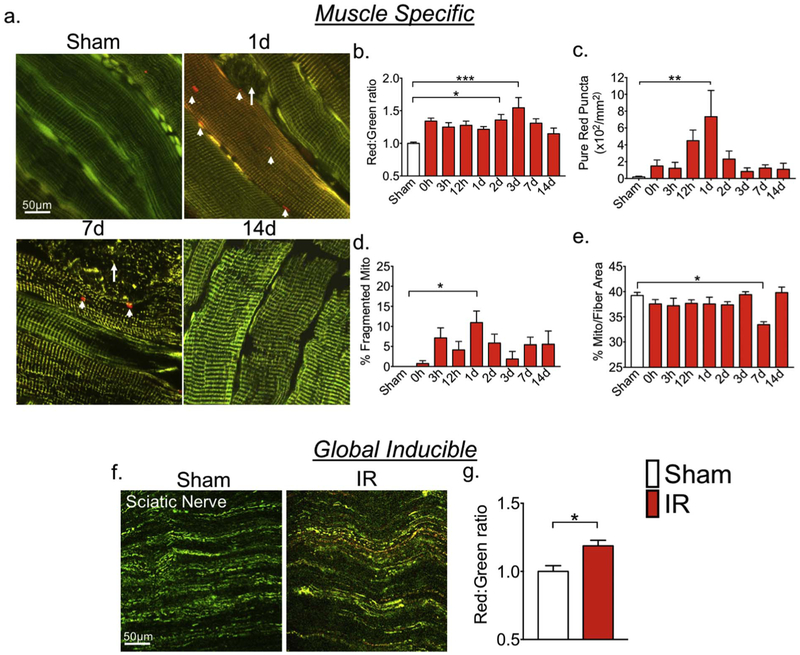
In order to gain information regarding the potential utility of CAG-CAT-MitoTimer mice, we began to characterize the response of muscle-specific MitoTimer mice to pathological stimuli. We subjected MCK-Cre;CAG-CAT-MitoTimer mice to tourniquet-induced IR injury, which is known to induce profound skeletal muscle dysfunction (Dorweiler et al., 2007; Lejay et al., 2014) as a result of excessive generation of mtROS and mitochondrial oxidative stress (Kalorgeris et al., 2012; Lejay et al., 2014). Consistent with this notion, we found that MitoTimer Red:Green ratio, which we have validated to be indicative of mitochondrial stress (Call et al., 2017; Laker et al., 2017; Laker et al., 2014), was significantly elevated at 2 and 3 days post-IR injury (Fig. 3a & b). The Red: Green ratio had a trend of increase at 0 h, but it was not statistically significant (p = 0.068). This may reflect feature of this reporter system for detecting cumulative changes. Pure red puncta, which are indicative of mitophagy (Call et al., 2017; Laker et al., 2017; Laker et al., 2014), were significantly increased 1 day post-IR injury (Fig. 3a & c) concurrent with a significant increase in fibers with fragmented mitochondria in nearly the whole myofiber (Fig. 3a & d). Interestingly, none of the fibers with fragmented mitochondria displayed pure red puncta, which are indicative of mitophagy. We speculate that the processes of massive mitochondrial fragmentation and mitochondrial oxidative stress may be exclusive of each other with the latter being part of apoptosis and/or necrosis. However, additional studies are necessary to determine the fate of these fibers (Lopes-ferreira et al., 2001; Grobler et al., 2004). In other words, the fibers containing pure red puncta may recover with ongoing mitophagy to remove damaged mitochondria caused by IR. Thus, mitochondrial fission could be a potential therapeutic target to preserve muscle fibers, as has been shown in the heart against IR injury (Gao et al., 2016). Finally, we measured mitochondrial content by quantifying MitoTimer positive pixels per fiber area (Laker et al., 2014) and showed a moderate, but significant reduction of mitochondrial content at 7 days following IR injury that was recovered by 14 days (Fig. 3a & d). Taken together, we show that muscle-specific MitoTimer mice could be a powerful tool for the simultaneous assessment of mitochondrial structure and content, oxidative stress, and mitophagy during lysosomal targeting in vivo.
Fig. 3.
MitoTimer reporter mice are sensitive for detection of mitochondrial changes in response to pathological stress in multiple tissues. MitoTimer transgenic mice were subjected to 1-h unilateral ischemia-reperfusion injury using a rubber band tourniquet followed confocal microscopy analysis. (a) Representative images of planatris muscles in muscle-specific MitoTimer mice at different time points over 14 days following IR injury. White arrows indicate fibers with a fragmented mitochondrial network, and arrow heads indicate pure red puncta. (b–e) Quantification of MitoTimer Red:Green ratio, pure red puncta, percent of fibers with mitochondrial network fragmentation, and mitochondrial content, respectively. *, **, *** denote p < 0.05, p < 0.01. p < 0.001, respectively; n = 5–7; (f) Representative images of MitoTimer signal in sciatic nerve in global inducible MitoTimer mice 3 h after ischemia-reperfusion (IR) comparing with the sham control (Sham); and (g) quantification of MitoTimer Red:Green ratio. * denotes p < 0.05. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To determine whether MitoTimer mice could be used for assessment of mitochondrial stress in other tissues, we subjected global inducible MitoTimer mice (CAG-CreERT2;CAG-CAT-MitoTimer) mice to IR following 7 days of daily injection of tamoxifen. We assessed MitoTimer Red:Green fluorescence 3 h following IR injury. IR led to a significant increase in MitoTimer Red:Green ratio in sciatic nerve exons (Fig. 3f and g) as well as in skeletal muscle (data not shown) compared with the sham control. As the network of mitochondria within the sciatic nerve was highly discontinuous under normal conditions (Fig. 3f), we did not observe further fragmentation, hence did not intend to quantify. These findings showed consistent behavior of MitoTimer protein between different tissues, although, the native state of the reticulum in a given tissue has to be considered when interpreting the observations of mitochondrial structure and quality.
3.4. Conclusion
Expanding from our established MitoTimer gene and its utilities in various model systems, we generated a conditional MitoTimer reporter mouse line. We confirmed tissue-specific expression of MitoTimer when crossing this mouse line into several tissue-specific Cre transgenic mouse lines, including inducible Cre lines. Using muscle-specific and global inducible MitoTimer mice, we showed that these reporter mice can be used for simultaneous measurements of mitochondrial structure, content, oxidative stress, and mitophagy at the step of lysosomal targeting of mitochondria-containing autophagosomes in skeletal muscle and mitochondrial oxidative stress in sciatic nerve. We have therefore obtained proof-of-principle evidence that CAG-CAT-MitoTimer mice, when crossed into Cre transgenic background, can be used to assess mitochondrial structure, oxidative stress and a specific step in mitophagy in any given tissue, and possibly, at any given time if using inducible form of Cre recombinase. This advancement of technology may help us gain unprecedented insight into mitochondrial structural and functional homeostasis.
Acknowledgments
This work was supported by NIH (R01-AR050429) to Z.Y. NIH (T32 HL007284-38) and ADA (1-16-PDF-030) to J.C.D. NIH (T32 HL007284-37) and AHA (114PRE20380254) to R.J.W. NIH (R01-DK062324) to M.D.O. NIH (R01-CA200755) to D.F.K. NIH (T32-DK0792922 and F32-DK108563) to H.M.P. The authors thank Volker Haase (Vanderbilt University) for providing PEPCK Cre mouse.
References
- Bruning JC, Michael MD, Winnay JN, Hayashi T, Accili D, Goodyear LJ, Kahn CR, 1998. A muscle-specific insulin receptor knockout exhibits features of the metabolic syndrome of NIDDM without altering glucose tolerance. Mol. Cell 2, 559–569. [DOI] [PubMed] [Google Scholar]
- Call JA, Wilson RJ, Laker RC, Zhang M, Kundu M, Yan Z, 2017. Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am. J. Phys. Cell Physiol 312, C724–C732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler B, Pruefer D, Andrasi TB, Maksan SM, Schmiedt W, Neufang A, Vahl CF, 2007. Ischemia-reperfusion injury pathophysiology and clinical implications. Eur. J. Trauma Emerg. Surg 600–612. 10.1007/s00068-007-7152-z. [DOI] [PubMed] [Google Scholar]
- Drake JC, Wilson RJ, Yan Z, 2015. Molecular mechanisms for mitochondrial adaptation to exercise training in skeletal muscle. FASEB J. 1–10. 10.1096/fj.15-276337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedlander SYG, Chu GC, Snyder EL, Girnius N, Dibelius G, Crowley D, Vasile E, Depinho RA, Jacks T, 2009. Article context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell 16, 379–389. 10.1016/j.ccr.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu A, Shi X, Zhang H, Fu B, 2017. Mitotherapy for fatty liver by intravenous administration of exogenous mitochondria in male mice. Front Pharmacol. 8, 1–8. 10.3389/fphar.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Yang J, Wu Y, Wang Q, Wang Q, Lai EY, Zhu J, 2016. Targeting dynamin 2 as a novel pathway to inhibit cardiomyocyte apoptosis following oxidative stress. Cell. Physiol. Biochem 39, 2121–2134. 10.1159/000447908. [DOI] [PubMed] [Google Scholar]
- Glancy B, Hartnell LM, Combs CA, Murphy E, Subramaniam S, Balaban RS, Glancy B, Hartnell LM, Combs CA, Fenmou A, Sun J, Murphy E, 2017. Power grid protection of the muscle mitochondrial power grid protection of the muscle mitochondrial reticulum. Cell Rep. 19, 487–496. http://dx.doi.Org/10.1016/j.celrep.2017.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobler LA, Collins M, Lambert MI, Sinclair-Smith C, Derman W, St Clair Gibson A, Noakes TD, 2004. Skeletal muscle pathology in endurance athletes with acquired training intolerance. Br. J. Sports Med 38, 697–704. 10.1136/bjsm.2003.006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins DF, Kimura K, Bernhardt WM, Shrimanker N, Akai Y, Hohenstein B, Saito Y, Johnson RS, Kretzler M, Cohen CD, Eckardt K, Iwano M, Haase VH, 2007. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J. Clin. Invest 117, 3810–3820. 10.1172/JCI30487.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodneland Nilsson LI, Nitschke Pettersen IK, Nikolaisen J, Micklem D, Avsnes Dale H, Vatne Røsland G, Lorens J, Tronstad KJ, 2015. A new live-cell reporter strategy to simultaneously monitor mitochondrial biogenesis and morphology. Sci. Rep 5, 17217. 10.1038/srep17217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju R, Wu W, Tang Q, Wu D, Xia Y, Wu J, 2015. Association analysis between the polymorphisms of HSD11B1 and H6PD and risk of polycystic ovary syndrome in Chinese population. PLoS One 10, 1–10. 10.1371/journal.pone.0140326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalorgeris T, Baines CP, Krenz M, Korthuis R, 2012. Cell biology of ischemia/reperfusion injury. Int. Rev. Cell Mol. Biol 298, 229–317. 10.1016/B978-0-12-394309-5.00006-7.Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuppertz L, Osiewacz H, 2016. Orchestrating the network of molecular pathways affecting aging: role of nonselective autophagy and mitophagy. Mech. Ageing Dev 153, 30–40. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, Levine B, 2010. Autophagy and the integrated stress response. Mol. Cell 40, 280–293. http://dx.doi.Org/10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, Zhang M, Royal MA, Hoehn KL, Driscoll M, Adler PN, Wessells RJ, Saucerman JJ, Yan Z, 2014. A novel mitotimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J. Biol. Chem 289, 12005–12015. 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laker RC, Drake JC, Wilson RJ, Lira VA, Lewellen BM, Ryall KA, Fisher CC, Zhang M, Saucerman JJ, Goodyear LJ, Kundu M, Yan Z, 2017. Ampk phosphorylation of Ulk1 is required for lysosome targeting of mitochondria in in exercise-induced mitophagy. Nat. Commun 8, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay A, Meyer A, Schlagowski A, Charles A, Bouitbir J, Pottecher J, Chakfé N, Zoll J, 2014. Mitochondria: mitochondrial participation in ischemia – reperfusion injury in skeletal muscle. Int. J. Biochem. Cell Biol 50, 101–105. 10.1016/j.biocel.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Lopes-ferreira M, Nu J, Rucavado A, Farsky SHP, Lomonte B, Angulo Y, Mari Â, Moura ANAM, Silva DA, 2001. Skeletal Muscle Necrosis and Regeneration after Injection of Thalassophryne nattereri (Niquim) Fish Venom in Mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I, Strobbe D, Frison M, Campanella M, 2015. Controlled and impaired mitochondrial quality in neurons: molecular physiology and prospective pharmacology. Pharmacol. Rec 99, 410–424. [DOI] [PubMed] [Google Scholar]
- Mcwilliams TG, Prescott AR, Allen GFG, Tamjar J, Munson MJ, Thomson C, Muqit MMK, Ganley IG, 2016. mito -QC illuminates mitophagy and mitochondrial architecture. In Vivo 214, 333–345. 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AH, Mccaffery JM, Chan DC, 2012. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis 50, 833–843. 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma LP, Duprez J, Takahashi HK, Gilon P, Wiederkehr A, Jonas J-C, 2012. Dynamic measurements of mitochondrial hydrogen peroxide concentration and glutathione redox state in rat pancreatic β-cells using ratiometric fluorescent proteins: confounding effects of pH with HyPer but not roGFP1. Biochem. J 441, 971–978. 10.1042/BJ20111770. [DOI] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstr??m KM, Fergusson MM, Yoo YH, Combs CA, Finkel T, 2015. Measuring in vivo mitophagy. Mol. Cell 60, 685–696. http://dx.doi.Org/10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terskikh A, 2000. “Fluorescent timer”: protein that changes color with time. Science 290, 1585–1588. 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- Toyama EQ, Herzig S, Courchet J, Lewis TL, Loson OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ, 2016. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281. 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez-Trincado C, García-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, Lavandero S, 2016. Mitochondrial dynamics, mitophagy and cardiovascular disease. J. Physiol. 594, 509–525. 10.1113/JP271301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov K, 2004. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem. Biol 11, 845–854. [DOI] [PubMed] [Google Scholar]
- Wanagat J, Hevener AL, 2016. Mitochondrial quality control in insulin resistance and diabetes. Curr. Opin. Genet. Dev. 38, 118–126. http://dx.doi.Org/10.1016/j.gde.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fang H, Groom L, Cheng A, Zhang W, Liu J, Wang X, Li K, Han P, Zheng M, Yin J, Wang W, Mattson MP, Kao JPY, Lakatta EG, Sheu SS, Ouyang K, Chen J, Dirksen RT, Cheng H, 2008. Superoxide flashes in single mitochondria. Cell 134, 279–290. http://dx.doi.Org/10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Tao C, Jiang L, Shao M, Ye R, Zhu Y, Gordillo R, Ali A, Lian Y, Holland WL, Gupta RK, Scherer PE, 2015. Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat. Cell Biol 17, 1099–1111. 10.1038/ncb3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AM, Asoh S, Ohsawa I, Ohta S, 2008. Imaging mitochondrial redox environment and oxidative stress using a redox-sensitive fluorescent protein. J. Nihon Med. Sch 75, 66–67. 10.1272/jnms.75.66. [DOI] [PubMed] [Google Scholar]
- Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ, 2001. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc. Natl. Acad. Sci. U. S. A. 98, 462–467. http://dx.doi.Org/10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]