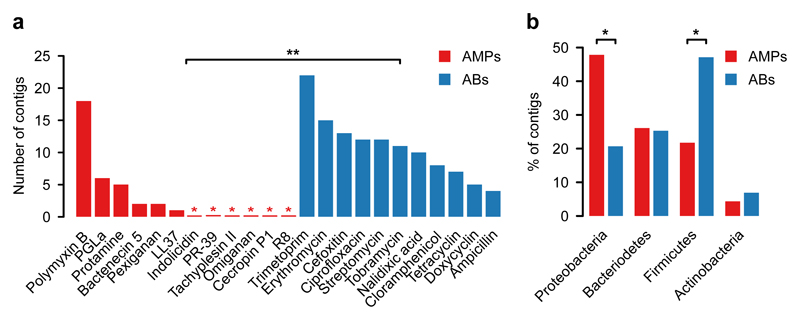
Fig. 2. In E. coli, short genomic fragments from the human gut microbiota confer AMP resistance (AMPs) less frequently than antibiotic resistance (ABs).
a) Functional selection of metagenomic libraries with 12 AMPs (red bars) resulted in fewer distinct resistance-conferring DNA contigs than with 11 conventional small-molecule antibiotics (ABs, blue bars). p=0.002 from two-sided negative binomial regression, n=34 (AMP resistance contigs), n=119 (AB resistance contigs). Red asterisks indicate zero values and ** indicates a significant difference between AMPs and antibiotics. b) Phylum-level distribution (%) of the AMP- (red bars) and antibiotic-resistance contigs (blue bars). In the case of AMPs, significantly more resistance contigs are originating from the Proteobacteria phylum (p=0.015, two-tailed Fisher's exact test, n=110), while contigs originating from the Firmicutes phylum are underrepresented (p=0.033, two-tailed Fisher's exact test, n=110). *Significant difference between AMPs and antibiotics for a given phylum.