Fig. 4. AMP resistance DNA fragments provide host-dependent phenotypic effects.
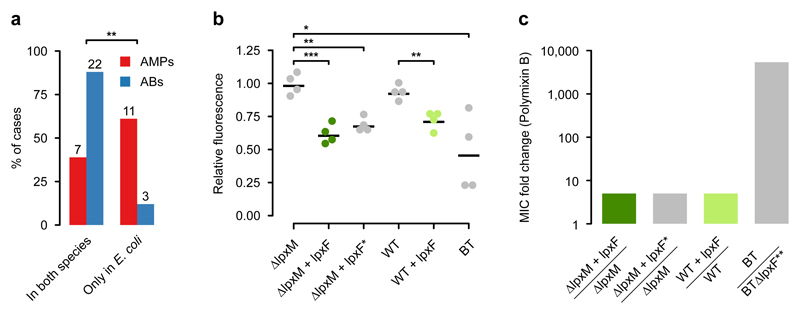
a) A significantly lower proportion of AMP resistance DNA fragments (AMPs, red bars) conferred resistance in both E. coli and S. enterica compared to antibiotic resistance DNA fragments (ABs, blue bars), suggesting weaker between-species conservation of the AMP resistance phenotypes. Asterisks indicate significant difference, p=0.0011, two-sided Fisher's exact test, n=16 for AMPs, n=25 for ABs. b) The IpxF ortholog from Parabacteroides merdae, isolated in our screen (marked as lpxFa here and in Table 1; represented with green colour) and a previously characterized fully functional IpxF from Francisella novicida35 (marked as lpxF*) decrease the net negative surface charge of ΔlpxM E. coli to a similar extent, and close to the level of wild-type Bacteroides thetaiotaomicron (BT) expressing its native lpxF. LpxFa has a similar effect both in ΔlpxM and wild-type E. coli (dark and light green dots). The fluorescence signal is proportional to the binding of the FITC-labeled poly-L-lysine polycationic molecules. Less poly-L-lysine binding reflects a less negative net cell surface charge36. *, **, *** indicate significant differences (p=0.03, 0.001 and 0.0004, respectively, Welch two-sample t-test, n=4 biological replicates, central horizontal bars represent mean values). Corresponding microscopic pictures are shown in Supplementary Fig. 11. c) LpxFa increases Polymyxin B resistance of both ΔlpxM and wild-type E. coli only five-fold (dark and light green bars) (n=3), to the same extent as LpxF from Francisella novicida (marked as lpxF*) (n=3). In contrast, lpxF in its original host, B. thetaiotaomicron (marked as lpxF**) provides a 5000-fold increment in Polymyxin B resistance12.