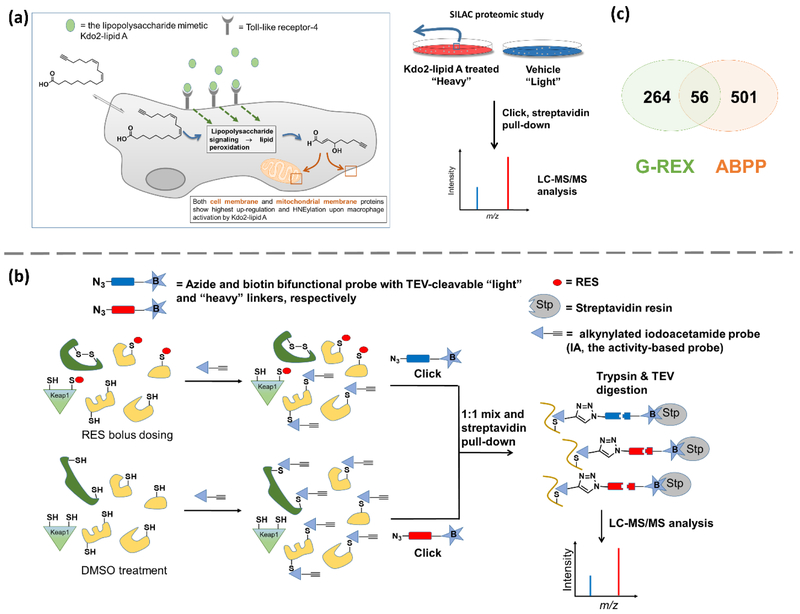
Figure 4.
(a) ω-Alkynyl linoleic acid incorporation for SILAC proteomics-based ID of RES-sensors [51]. Cells fed with terminal alkyne-labeled linoleic acid are activated with Kdo2-lipidA to produce terminal alkyne-labeled RES through lipid peroxidation. Proteins labeled under these conditions are identified through SILAC proteomics analysis. Plasma and mitochondrial membrane proteins are the most prevalent HNEylated targets identified under these conditions. (b) Competitive isoTOP-ABPP of RES-modified proteins[22]. Parallel sets of intact cells/lysates are treated with RES or DMSO. After cell lysis (where applicable), both sets of lysates are incubated with a reporter electrophile [e.g., iodoacetamide (IA)] which non-specifically labels (a subset of) cysteines. Using biotin-azide tags (housing differential isotope-labeled linker bearing TEV-protease cleavage site in between azide and biotin), Click coupling biotinylates the IA-labeled cysteines. Steptavidin pull-down followed by trypsin digest and TEV cleavage allow for LC-MS/MS analysis that indirectly identifies RES-modified cysteines based on loss of IA-labeled cysteine signals. Care must be exercised to exclude potential false negatives/positives. For instance, proteins (in dark green) subject to allosteric regulation, disulfide modification, and/or down-regulation in response to RES-bulk exposure, may be falsely identified as direct HNE-interactors. Proteins, [e.g., Keap1 (in light green)] with reactive cysteines with preference to undergo conjugate addition reaction to sp2-derived RES such as HNE, may not react extensively with sp3–centered electrophilic reporter probes (such as IA). Some proteins especially those unstable to oxidation-induced aggregation/precipitation may not be stable to sample processing conditions prior to MS analysis. (c) Venn diagram showing overlap of HNE-targeted proteins detected by G-REX (top 300)[17] and ABPP (all probe-labeled proteins)[22]. Note: the data originate from HEK293T and MDA-MB 231 cells in G-REX and ABPP, respectively.