FIGURE 1.
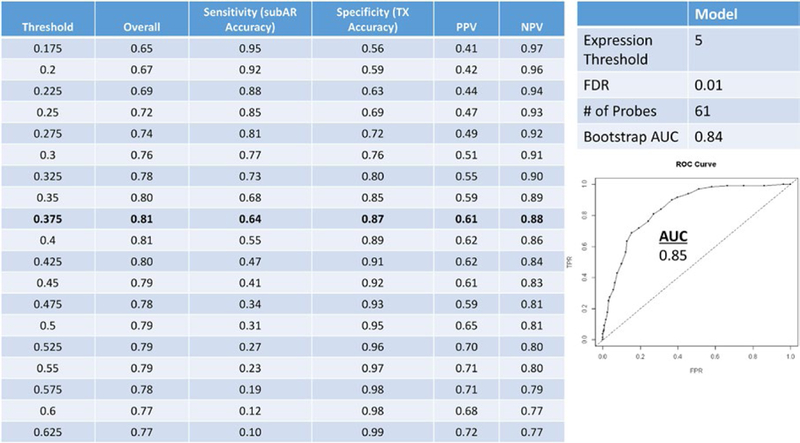
Discovery on 530 Clinical Trials in Organ Transplantation 08 (CTOT‐08) paired peripheral blood and surveillance biopsy samples cohort. We ran the top random forests model from the differentially expressed gene data with threshold selection and predictive metrics on 530 paired samples from the CTOT‐08 (400 [75.5%] transplant excellent [TX]; 130 [24.5%] subclinical acute rejection [subAR]) discovery training set cohort (100000 trees, expression threshold of 5, and false discovery rate [FDR] 0.01), optimizing for area under the curve (AUC; 0.85 [0.84 after internal validation by resampling bootstrap]). A predicted probability threshold of 0.375 was selected, yielding an overall accuracy of 0.81, specificity and negative predictive value (NPV) (87% and 88%, respectively) over sensitivity, and positive predictive value (PPV) (64% and 61%, respectively). The classifiers consisted of 61 probe sets mapping to 57 genes
