SUMMARY
Triple-negative breast cancer (TNBC) remains the most challenging breast cancer subtype to treat. To date, therapies directed to specific molecular targets have rarely achieved clinically meaningful improvements in outcomes of patients with TNBC, and chemotherapy remains the standard-of-care. Here we seek to review the most recent efforts to classify TNBC based on comprehensive profiling of tumors for cellular composition and molecular features. Technological advances allow for tumor characterization at ever increasing depth, generating data that, if integrated with clinical-pathologic features, may help improve risk stratification of patients, guide treatment decisions and surveillance, and help identify new targets for drug development.
INTRODUCTION
Breast cancer is the most frequently diagnosed cancer and the second most common cause of cancer mortality in women worldwide (1). Breast tumors that are immunohistochemically characterized by lack of estrogen receptor (ER), progesterone receptor (PR), and HER2 (also defined by lack of HER2 amplification by FISH) are classified as triple-negative breast cancer (TNBC) and account for approximately 15–20% of all breast carcinomas (2). Compared to hormone receptor-positive or HER2-positive disease, TNBC has a highly aggressive clinical course, with earlier age of onset, greater metastatic potential, and poorer clinical outcomes as shown by the higher relapse and lower survival rates (2,3). The molecular mechanisms that drive TNBC recurrence have not been fully elucidated. Consequently, to date, targeted therapies have not significantly improved survival in TNBC patients, and chemotherapy remains the standard-of-care. Although many patients with early stages of TNBC are cured with chemotherapy, in those who develop metastatic disease, median OS (overall survival) with current treatment options is 13–18 months (4).
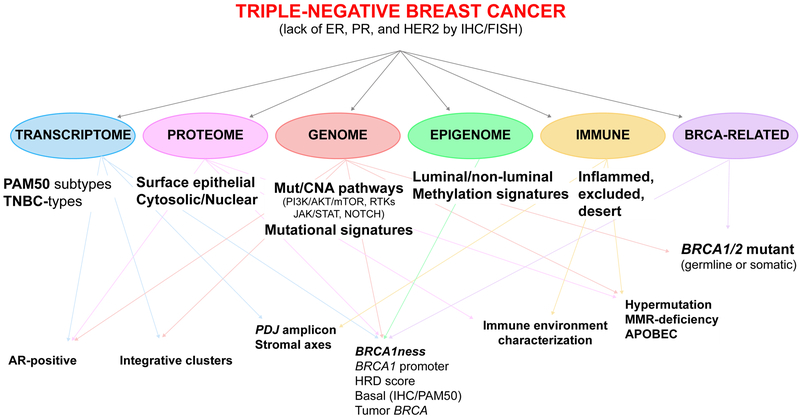
Major effort has been devoted over the past decade to classify TNBC into distinct clinical and molecular subtypes that could guide treatment decisions. Characterization of genomic, transcriptomic, proteomic, epigenomic, and microenvironmental alterations have expanded our knowledge of TNBC. Here we review the most recent innovations in TNBC molecular taxonomy, the complex interaction between these classifications (Figure 1), and their potential therapeutic implications.
Figure 1.
Overview of the complex interactions between molecular classifications of TNBC based on genomic, transcriptomic, proteomic, epigenomic and immune characterization of the tumor and its microenvironment. ER: estrogen receptor; PR: progesterone receptor; CNA: copy number alterations; AR: androgen receptor; HRD: homologous recombination deficiency; IHC: immunohistochemistry; TIL: tumor-infiltrating lymphocytes.
TNBC and intrinsic breast cancer subtypes
Early transcriptomic profiling of breast cancer using microarrays classified tumors into five intrinsic subtypes: luminal A, luminal-B, HER2-enriched, basal-like, and a normal breast-like group (5,6). Although all intrinsic subtypes can be found within immunohistochemically (IHC)-defined triple-negative disease, basal-like tumors exhibit the greatest overlap with TNBC. Between 50–75% of TNBC have basal phenotype and approximately 80% of basal-like tumors are ER-negative/HER2-negative (Figure 2) (7,8). Characterization of intrinsic subtypes using a 50-gene assay (established as the PAM50 subtype predictor) has provided independent predictive information of pathologic complete response (pCR) to neoadjuvant therapy across all subtypes (9), but when restricting analyses to TNBC, none of the PAM50 signatures at time of diagnosis have significantly correlated with pCR (10). In basal-like TNBC, low expression of the luminal-A signature and high expression of the proliferation score were both significantly associated with pCR (10). High expression of cell cycle-related (e.g., CCNE, FANCA) and low levels of estrogen signaling-related (e.g., FOXA1, PGR) genes were associated with pCR, while high expression of epithelial-mesenchymal transition (EMT) genes (e.g., TWIST1, ZEB1) was significantly enriched in residual disease (10). Again, in the adjuvant setting, no significant gene-signature predictors of disease-free survival (DFS) have been found in TNBC (10). However, in basal-like TNBC in GEICAM/9906, and in basal-like tumors treated with adjuvant chemotherapy in the METABRIC dataset and in CALGB/9741, the two previously identified signatures (low luminal-A, high proliferation score) predicted improved DFS and recurrence-free survival (RFS).
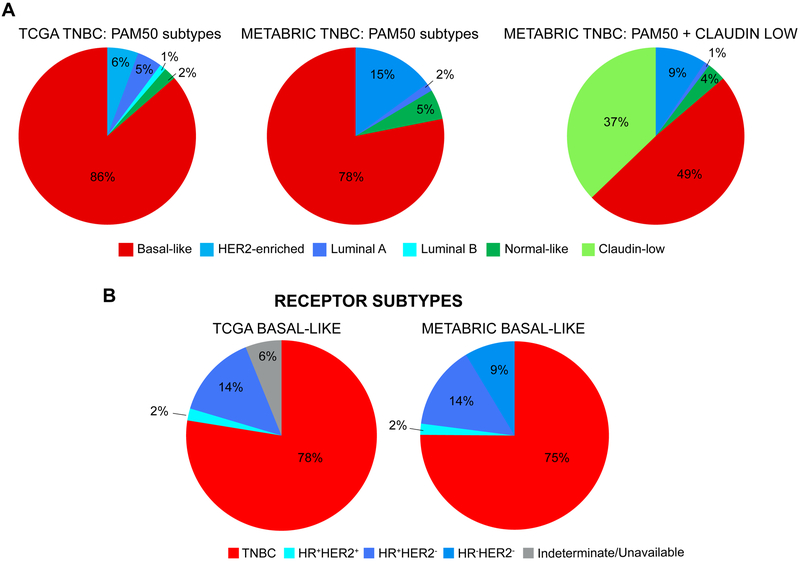
Figure 2.
Distribution of intrinsic subtypes among TNBC and distribution of TNBC among basal-like breast cancer. A, Comparison of distribution of intrinsic subtypes defined by PAM50 and PAM50+Claudin-low in TCGA and METABRIC datasets in triple-negative breast cancer (TNBC). TNBC was defined as clinical ER, PR and HER2 negative testing per IHC. In TCGA, 88 TNBC samples had available PAM50 data. The distribution of intrinsic subtypes was: basal-like (86%), HER2-enriched (6%), luminal-A (5%), luminal-B (1%), and normal-like (2%). In METABRIC, 320 TNBC samples had available intrinsic subtype data. When including claudin-low in the PAM50 predictor, the distribution of subtypes was: basal-like (49%), claudin-low (37%), HER2-enriched (9%), normal-like (4%), luminal-A (1%), and luminal-B (0%). When excluding the 119 samples with claudin-low subtype, the distribution of subtypes was: basal-like (78%), HER2-enriched (15%), normal-like (5%), luminal-A (2%), and luminal-B (0%). B, Comparison of distribution of breast cancer subtype according to receptor status defined by IHC in TCGA and METABRIC datasets in basal-like breast cancer. Of 98 basal-like breast cancers in TCGA, 78% were TNBC per IHC. Of 209 basal-like breast cancers (PAM50+Claudin-low classifier) in METABRIC, 75% were TNBC. Figures generated by re-analysis of publicly available (22,36,37) using cBioPortal (150,151).
PAM50-defined subtypes have not yet been validated as predictors of benefit to individual chemotherapeutic agents in TNBC. An increase in pCR rates from 47% to 61% was noted with the addition of carboplatin to neoadjuvant therapy in patients with basal-like TNBC in CALGB/40603 (11), although this improvement did not differ from that observed in the overall population after incorporating the small number of non-basal-like tumors. In the metastatic setting, carboplatin and docetaxel achieved comparable objective response rates (ORR) in basal-like tumors in the TNT trial (32.5% vs. 31.0%, respectively; p=0.87) (12). Of note, though a significant interaction was observed between PAM50 subgroups and treatment arm, this was driven by the unexpected finding of greater efficacy of docetaxel compared to carboplatin in non-basal-like tumors (ORR 72.2% vs. 16.7%; p=0.002) (12). Further studies prospectively evaluating taxanes and other agents in predefined subgroups are needed to confirm any differential activity in non-basal-like TNBC.
Additional gene expression analyses later revealed the presence of another intrinsic subtype, claudin-low, present in 7–14% of all breast cancers (6). Approximately 70% of claudin-low tumors are TNBC, with high representation of metaplastic and medullary breast carcinomas. While claudin-low and basal-like subtypes share low luminal and HER2 gene expression, claudin-low tumors do not highly express proliferation genes. They are uniquely characterized by low levels of cell adhesion proteins and elevated expression of immune-related genes (e.g., CD4, CD79a). These mesenchymal features (including elevated expression of CD44, vimentin, N-cadherin) and low epithelial differentiation (low CD24 gene expression) resemble a mammary stem cell-like phenotype (CD44+CD24−/low) that can be acquired by EMT (6). In retrospectives studies, claudin-low tumors were associated with lower (39%) pCR rates compared to basal-like subtype (73%), and worse prognosis than luminal-A tumors but similar survival as luminal-B, HER2-enriched, or basal-like tumors (6). Formation of cancer stem cells is induced by TGFβ in claudin-low cell lines (13), and in chemotherapy-resistant TNBC, TGFβ signaling and other stem cell markers are overexpressed (14). Thus, inhibition of TGFβ signaling may represent a potential therapeutic strategy to help prevent the development of chemo-refractory disease, particularly in the claudin-low subtype.
Molecular definition of TNBC heterogeneity
With evolving transcriptomic studies, the heterogeneity of TNBC has been further dissected. Lehmann et al. analyzed 21 public microarray datasets filtered for TNBC based on ER, PR, and HER2 expression, and identified seven clusters within TNBC: basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal-stem-like (MSL), luminal androgen receptor (LAR), and an unstable cluster (UNS) (15). These subtypes are characterized by distinct patterns of molecular alterations, both in terms of RNA expression, somatic mutations and copy number variations, that tend to cluster in genes implicated in specific pathways. The BL1 subtype, enriched in genes involved in DNA damage response and cell-cycle regulation (including the highest rate of TP53 mutations [92%], high gain/amplifications of MYC, CDK6 or CCNE1, and deletions in BRCA2, PTEN, MDM2 and RB1) (16) and the BL2 subtype, with high levels of growth factor signaling and metabolic pathway activity, share a highly proliferative phenotype that correlates with improved pCR with mitotic inhibitors, such as taxanes. Genes involved in antigen processing and presentation, immune cell and cytokine signaling (e.g., JAK/STAT, TNF, NFKB) pathways are highly expressed in the IM subtype. Mesenchymal-like TNBC subtypes, M and MSL, display similar expression profiles related to cell motility, differentiation and EMT, but are discernible by the unique enrichment in MSL of angiogenesis- and stem cell-associated genes, and low claudin expression. Finally, despite ER-negativity, the LAR subtype displays a luminal pattern of gene expression (e.g., high levels of FOXA1, GATA3, SPEDF, and XBP1), with elevated mRNA and protein levels of AR, overlapping in 82% of cases with luminal-A or luminal-B intrinsic subtypes. Thus, not surprisingly, LAR tumors are enriched in mutations in PIK3CA (55%), KMT2C (19%), CDH1 (13%, in conjunction with a higher prevalence of invasive lobular histology), NF1 (13%), and AKT1 (13%) (16). The 7-subtype classification independently predicted pCR, but not distant metastasis-free or overall survival (OS) in a retrospective analysis of patients with TNBC treated with neoadjuvant chemotherapy (17). Median OS was highest in LAR and BL1 subtypes, despite low pCR rate in the LAR group. Follow-up in vitro studies with representative cell lines of TNBC subtypes demonstrated differential drug sensitivity that, if validated, may have clinically relevant implications (15). Of note, all seven clusters were not detected in an independent analysis of 5 datasets of IHC-identified TNBC, as opposed to gene expression-defined TNBC (15). Even across other studies in which TNBC was identified using mRNA expression, reproducibility of the BL2 and UNS subtypes has not been consistent (16,17).
In a follow up study, by performing histological assessment and laser microdissection prior to RNA isolation and gene expression analysis, Lehmann and colleagues confirmed that the presence of stromal cells in tumor specimens – such as infiltrating lymphocytes and tumor-associated mesenchymal cells – influences the definition of IM and MSL subtypes, respectively (18). This led to a revised classification, TNBCtype4, into four stable transcriptional subtypes (BL1, BL2, M, and LAR) that significantly differ not only in prognosis and response to chemotherapy, but also in initial presentation and patterns of recurrence, where regional nodal involvement is more common in LAR TNBC and metastatic recurrences have tropism to the lung in M subtypes and to the bone in LAR subtypes. Similarly to the 7-subtype classification, response to neoadjuvant chemotherapy (platinum- and taxane-based regimen) is significantly associated with TNBCtype4 subtypes (p=0.027), with the highest and lowest pCR rates reported in BL1 (65.6%) and LAR (21.4%), respectively (19). These findings highlight a major limitation of classifiers defined based on profiling of bulk tumors that cannot distinguish between tumor and stromal cells and support the increasing use of single-cell techniques to improve the characterization of the tumor and its microenvironment. In fact, single-cell RNA sequencing has demonstrated the presence of multiple subtypes within most primary TNBC tumors, suggesting that the dominant signature identified through bulk sequencing may not accurately inform underlying biological processes, including interactions between malignant and normal stromal cell types (20). Differences in the prevalence of intratumoral heterogeneity between TNBC and ER-positive breast cancer could partly explain the challenges to date to apply commercially available gene expression assays in routine clinical practice to provide prognostic and predictive information in TNBC.
Additional efforts to distinguish stable molecular TNBC phenotypes using gene expression profiling include the classification into four subtypes by Burstein et al.: LAR, Mesenchymal (MES), Basal-like Immune Suppressed (BLIS), and Basal-like Immune Activated (BLIA) (21). Interestingly, the BLIS subtype exhibited the worst prognosis and the BLIA subgroup conferred the best outcome in terms of disease-free survival. In addition, specific DNA copy number variations were identified in each subtype, such as focal gains on 11q13 (CCND1, FGF family) in the LAR subtype or BLIA-specific overexpression of CTLA4. In another analysis that integrated somatic copy number variations and gene expression profiles of primary breast tumors of any IHC-subtype in the METABRIC dataset, 10 integrative clusters were identified, where IntClust 10 exhibited the greatest overlap with PAM50 basal-like tumors and was characterized by 5 loss/8q gain/10p gain/12p gain (22). As exemplified by studies assessing the overlap between these different gene expression classifications, a high correlation has been described between PAM50-defined basal-like, Lehmann BL1/BL2 and Baylor BLIA/BLIS subtypes (21–23), emphasizing the high stability of the basal subtype across TNBC. These studies also highlight the inherent problems associated with the TNBC definition, since it does not reflect a clear molecular entity. What seems clear is that luminal (ER-positive or AR-positive) and non-luminal (basal and mesenchymal) tumors have very different evolutionary paths, and this is in part likely driven by their normal cell-of-origin reflected in distinct epigenetic profiles. Thus, improved classifications based on epigenetic profiles and quantitative measures of intratumoral heterogeneity may lead to a better definition of clinically relevant TNBC subtypes.
Androgen receptor-positive TNBC
As detailed above, a luminal phenotype, characterized by expression of the androgen receptor (AR) and luminal lineage-driving transcription factors, has been consistently identified across several studies in TNBC. In Core-Basal tumors, the prevalence of AR-positivity defined by ≥1% of tumor cell nuclei IHC staining has been reported to be 32% (24). Interestingly, other studies have suggested that LAR tumors are characterized by a quiescent cell state (25), as opposed to rapidly proliferative basal tumors, raising the question of the optimal method of testing for AR positivity and possibly lack of a robust approach due to limited sample size. Altogether, this has prompted interest in exploring the role of anti-androgens in this subgroup. In vivo studies have shown that tumors derived from LAR cell lines (e.g., MDA-MB-453, SUM185PE, CAL-148) are highly sensitive to the AR antagonist, bicalutamide (15). In phase II single-arm trials conducted in patients with metastatic AR-positive, ER/PR-negative breast cancer, bicalutamide and enzalutamide demonstrated stable disease at 6 months of 19% and 28%, respectively, though no objective responses were observed (26,27). Abiraterone acetate and prednisone achieved a similar 20% clinical benefit rate (CBR) at 6 months, and although the study failed to meet the prespecified >25% cutoff necessary to reject the null hypothesis, prolonged responses were observed (range: 6.4–23.4 months) (28). An androgen-driven genomic signature, Dx, predicted improved OS with enzalutamide (29), and this led to the design of a phase III trial comparing enzalutamide, paclitaxel and the combination in selected Dx-positive advanced TNBC (NCT02929576).
Similar to luminal tumors, strategies to enhance the effectiveness of hormone receptor blockade have been pursued in AR+ TNBC. Enrichment in PIK3CA mutations has been described in triple-negative tumors that are AR+ (36–40%) by IHC compared to AR− (4–9%) (30,31), the majority of which are located in the kinase domain H1047 mutational hotspot and co-occur with amplification of the PIK3CA locus (30). Combination of PI3K/mTOR inhibition and AR antagonism has demonstrated synergistic activity in AR+ TNBC preclinical models, and a phase I trial is planned to explore enzalutamide plus alpelisib, an α-specific PI3K inhibitor, in patients with AR+, PTENlow (IHC 0%) TNBC (NCT03207529). Additional studies have revealed that, in contrast to basal-like and mesenchymal subtypes, LAR TNBC cell lines are highly sensitive to CDK4/6 inhibitors, with comparable sensitivity to that observed in the ER+ MCF7 cell line (25). LAR cell lines exhibit lower transcriptomic levels of CCNE1 and CDK2 compared to basal-like TNBC and, thus, are dependent on CDK4/6 to phosphorylate RB1 and re-enter the cell cycle. In vitro PI3K inhibition decreases post-mitotic CDK2 activity in PIK3CA-mutant TNBC, suggesting potential sensitization to CDK4/6 inhibitors, including in non-LAR TNBC (25); this has provided the rationale for the ongoing clinical trial testing palbociclib combined with either taselisib or pictilisib in PIK3CA-mutant ER+− breast cancer (NCT02389842).
Protein markers in TNBC for targeted antibody-drug conjugates
Isolation of glycoproteins on the surface of epithelial cancer cells has triggered the development of antibody-drug conjugates (ADC) designed to improve delivery of elevated concentrations of cytotoxic drugs to cells expressing these molecules. Many of these targets are not necessarily cancer drivers or specific to breast cancer; instead, they require differential protein expression in malignant versus normal cells. Interestingly, several ADC have demonstrated encouraging activity in TNBC. Sacituzumab govitecan (IMMU-132) is an antibody-SN-38 conjugate targeting Trop-2, which is expressed in almost 90% of TNBC (32). In patients with heavily pre-treated metastatic TNBC, IMMU-132 achieved an ORR of 30%, and median PFS and OS were 6.0 and 16.6 months, respectively. LIV-1 is a transmembrane protein with metalloprotease activity expressed in 68% of metastatic TNBC samples. Ladiratuzumab vedotin (SGN-LIV1A), with monomethyl-auristatin-E (MMAE) as the payload, yielded a 25% ORR in a similar population of patients with TNBC, and median PFS was 11 months (33). Significant expression of glycoprotein-NMB (gpNMB), defined as staining ≥25% of tumor epithelial cells, is present in approximately 40% of TNBC, and in this subgroup, glembatumumab vedotin (CDX-011, an ADC that binds to gpNMB to deliver MMAE) achieved 40% ORR versus 0% with investigator´s choice of therapy (34). However, when compared to capecitabine in preselected gpNMB-overexpressing metastatic TNBC in the METRIC phase II trial, glembatumumab vedotin failed to demonstrate improved PFS, ORR or OS, leading to discontinuation of the development of this ADC (Celldex’s METRIC Study Press release April 16, 2018; https://globenewswire.com/news-release/2018/04/16/1471890/0/en/Celldex-s-METRIC-Study-in-Metastatic-Triple-negative-Breast-Cancer-Does-Not-Meet-Primary-Endpoint.html). SGN-LIV1A is currently being evaluated in phase II trials, and IMMU-132 has advanced to phase III development (ASCENT: NCT02574455). Given the high prevalence of many of these markers in TNBC, IHC confirmation may not be necessary prior to starting therapy, but other proteins overexpressed less frequently may require prescreening efforts to help identify patients who are more likely to benefit from ADC.
Somatic genetic alterations in TNBC
Cancers harbor numerous somatic genetic alterations, though only a small proportion of them confer clear fitness advantage, also known as ¨cancer drivers¨ (35). Large-scale exome and targeted sequencing studies in primary breast tumors have revealed the presence of many alterations in putative cancer driver genes in TNBC (36–38). The average mutation rate in basal-like breast cancer is among the highest in breast tumors, 1.68 mutations per megabase (Mb); tumors that reach rates greater than three standard deviations above the mean (>4.68 mutations/Mb) are considered hypermutated (36). Different genomic classifications in breast cancer have been proposed by grouping NGS-detected alterations in known cancer-driver genes according to the intracellular pathways in which they are involved, such as PI3K/AKT and RAS/MAPK signaling, DNA damage repair, and cell cycle or transcriptional regulation (Table 1) (36,37,39).
Table 1.
Classifications according to potentially targetable pathways based on exome or targeted sequencing
| TCGA (Basal-like) (36) | Genomic alteration (frequency, %) |
|---|---|
| TP53 pathway | TP53 mut (84); gain of MDM2 (14) |
| PIK3CA/PTEN pathway | PTEN mut/loss (35); INPP4B loss (30); PIK3CA mut (7) |
| RB1 pathway | RB1 mut/loss (20); CCNE1 amp (9); high expression of CDKN2A; low RB1 expression |
| METABRIC (ER-negative) (37) | Mutated gene (frequency, %) |
|---|---|
| AKT signaling | PIK3CA (24), AKT1 (2), PTEN (4), PIK3R1 (3), FOXO3 (1) |
| Cell cycle regulation | RB1 (4), CDKN2A (1) |
| Chromatin function | KMT2C (9), ARID1A (3), NCOR1 (2), PBRM1 (3), KDM6A (2) |
| DNA damage and apoptosis | TP53 (77), BRCA1 (3), BRCA2 (3) |
| MAPK signaling | NF1 (4), MAP3K1 (3), MAP2K4 (1), KRAS (1) |
| Tissue organization | CDH1 (3), MLLT4 (3) |
| Transcription regulation | TBX3 (2), RUNX1 (2), GATA3 (1), ZFP36L1 (1), MEN1 (1) |
| Ubiquitination | USP9X (3), BAP1 (3) |
| Other | ERBB2 (3), SMAD4 (1), AGTR2 (1) |
| Residual disease post-neoadjuvant chemotherapy (Triple-negative) (39) | Genomic alteration (frequency, %) |
|---|---|
| Cell cycle | RB1 loss (11); CDKN2A loss (9); CDKN2B loss; CDK4 amp; CDK6 amp (6); CCND1 amp (6); CCND2 amp (6); CCND3 amp (6); CCNE1 amp (6); AURKA amp |
| PI3K/mTOR pathway | PTEN mut/loss (16); PIK3CA mut/amp (12); PIK3R1 mut/amp; AKT1 amp; AKT2 amp; AKT3 amp (7), RAPTOR amp; RICTOR amp; TSC1 truncations/mut |
| Growth factor receptor | IGF1R amp (6); EGFR amp (4); MET amp; KIT amp; FGFR1 amp; FGFR2 amp; FGFR4 amp |
| RAS/MAPK pathway | KRAS amp/gain (7); BRAF amp/gain; RAF1 amp/gain; NF1 truncations (7) |
| DNA repair | BRCA1 truncations/loss/mut (11); BRCA2 truncations/loss/mut; ATM mut |
| JAK2/STAT3 pathway | JAK2 amp (10) |
Mut: gene mutation; Gain: gene copy number gain (<5 but more than 2 copies); Amp: gene amplification (≥5 copies and/or gene-specific and centromeric probe ratio >2). The definition of copy number gain vs. amplification is somewhat platform and study dependent. In general, copy number gain ≥5 is considered an amplification, while copy number gain >2 but below 5 is considered a copy number gain. But, some studies define amplification when gene-specific vs. centromeric probe ratio is >2. Frequencies (%) of alterations are included when available.
Most somatic mutations in TNBC occur in tumor suppressor genes (e.g., TP53, RB1, PTEN), which have not been successfully targeted therapeutically to date. Although less prevalent, oncogenic alterations in the PI3K/AKT pathway have also been described in basal-like breast cancer (PIK3CA mutation, 7%; AKT3 amplification, 28%; PTEN mutation or loss, 35%) (36), potentially qualifying patients for clinical trials with matched therapies. Consistent with findings in untreated triple-negative tumors, targeted sequencing of residual disease post-neoadjuvant chemotherapy showed that >90% of patients had at least one altered pathway (39). However, only three alterations were found to be significantly prognostic for OS (JAK2 amplification, BRCA1 truncation or mutation: predicted poor OS; PTEN alteration: better OS). Drugs that inhibit these pathways have been explored in clinical trials in TNBC, mostly in combination with other therapies due to limited single-agent activity (Table 2).
Table 2.
Efficacy of Genomic-Based Targeted Therapies in Clinical Trials in Triple-Negative Breast Cancer.
| Pathway | Drug | Mechanism | Patient population |
Trial design (Total N patients) |
Intervention | Exploratory biomarker |
Efficacy |
Clinicaltrials.gov
Identifier |
|---|---|---|---|---|---|---|---|---|
| PI3K/AKT/mTOR | Buparlisib | PI3K inhibitor | Locally advanced/metastatic HER2-negative | Randomized phase II (n=416) (43) | Buparlisib + Paclitaxel vs. Placebo + Paclitaxel |
Stratification by PI3K pathway activation | PFS (full population): 8.0 vs. 9.2 (HR 1.18; 95% CI: 0.82-1.68) | NCT01572727 |
| PFS (PI3K-activated): 9.1 vs. 9.2 (HR 1.17; 95% CI: 0.63-2.17) | ||||||||
| PFS (TNBC): 5.5 vs. 9.3 (HR 1.86; 95% CI: 0.91-3.79) | ||||||||
| Ipatasertib | AKT inhibitor | Locally advanced/metastatic TNBC | Randomized phase II (n=124) (44) | Ipatasertib + Paclitaxel vs. Placebo + Paclitaxel |
Stratification by tumor PTEN status | PFS (intent-to-treat): 6.2 vs. 4.9 (HR 0.60; 95% CI: 0.37-0.98; p=0.037) | NCT02162719 | |
| PFS (PTEN-low): 6.2 vs. 3.7 (HR 0.59; 95% CI: 0.26-1.32; p=0.18) | ||||||||
| PFS (PIK3CA/AKT1/PTEN-altered): 9.0 vs. 4.9 (HR 0.44; 95% CI: 0.20-0.99; p=0.041) | ||||||||
| MK2206 | AKT inhibitor | Neoadjuvant stage II-III breast cancer (any subtype) | Randomized phase II (n=149) (45) | Paclitaxel +/− MK2206 (followed by AC) | NA | pCR (all): 35.2 vs. 21.1 | NCT01042379 | |
| pCR (TNBC): 40.2 vs. 22.4 | ||||||||
| Temsirolimus, Everolimus | mTORC1 inhibitor | Metastatic metaplastic TNBC | Phase I dose expansion (n=52) (46) | Liposomal doxorubicin + Bevacizumab + (Temsirolimus or Everolimus) | Exploratory analysis by PI3K pathway activation | ORR (all): 21 (95% CI: 11-35) | NCT00761644 | |
| ORR (PI3K-activated): 31 (95% CI: 16-50) | ||||||||
| Everolimus | mTORC1 inhibitor | Neoadjuvant stage II-III TNBC | Randomized phase II (n=145) (47) | Cisplatin + Paclitaxel + Everolimus vs. Cisplatin + Paclitaxel + Placebo |
Exploratory analysis of mutated genes, TNBC subtype, Ki67, AR and TILs | pCR (all): 36 vs. 48 (p=0.41) | NCT00930930 | |
| EGFR | Panitumumab | EGFR monoclonal antibody | Locally advanced/metastatic TNBC | Non-randomized phase II (n=71) (49) | Panitumumab + Carboplatin + Gemcitabine | EGFR amp, p53 loss, PTEN loss, PIK3CA mut | PFS (all): 4.4 (95% CI: 3.2-5.5) | NCT00894504 |
| PFS (EGFR-amp): 3.42 (95% CI: 1.51-NR) | ||||||||
| Cetuximab | EGFR monoclonal antibody | Neoadjuvant stage II-IIIA TNBC | Non-randomized phase II (n=28) (50) | Cetuximab + Docetaxel | EGFR, Ki67, Cytokeratins, CD8/FOXP3 | pCR (intent-to-treat): 25 (95% CI: 9-41) | NCT00600249 | |
| Lapatinib | EGFR/HER2 inhibitor | Locally advanced/metastatic HER2-negative | Randomized phase III (n=580) (51,149) | Lapatinib + Paclitaxel vs. Placebo + Paclitaxel |
EGFR | EFS (TNBC): 4.6 vs. 4.8 (HR: 1.25; 95% CI: 0.85-1.83) EFS (TNBC EGFR+): 4.2 vs. 4.9 EFS (TNBC EGFR−): 5.2 vs. 4.3 |
NCT00075270 | |
| RAS/MAPK | Cobimetinib | MEK1/2 inhibitor | Locally advanced/metastatic TNBC | Open-label safety run-in (n=16), randomized phase II (n=90) (55) | Cobimetinib + Paclitaxel vs. Placebo + Paclitaxel |
TNBC subtype, genetic alterations, PD-L1 expression | PFS (intent-to-treat): 5.5 vs. 3.8 (HR 0.73; 95% CI: 0.43-1.24; p=0.25) | NCT02322814 |
| JAK/STAT | Ruxolitinib | JAK1/2 inhibitor | Metastatic TNBC or IBC of any subtype | Non-randomized phase II (n=21) (66) | Ruxolitinib | JAK2 amplification, pSTAT3 | PFS (all): 1.2 (95% CI: 0.97-1.84) | NCT01562873 |
| NOTCH | PF-03084014 | Gamma-secretase inhibitor | Metastatic HER2-negative breast cancer | Phase I dose-finding/dose-expansion (n=29) (67) | PF-03084014 + Docetaxel | NA | ORR: 16 (95% CI: 4.5-36.1) | NCT01876251 |
Main efficacy analyses of biomarker-selected subgroups of interest are highlighted. HR, 95% CI and p values are included when available. TNBC: triple-negative breast cancer; PFS: progression-free survival (months); pCR: pathologic complete response (%); ORR: objective response rate (%); IBC: inflammatory breast cancer; AR: androgen receptor; AC: adriamycin/cyclophosphamide; HR: hazard ratio; CI: confidence interval; N: number; NA: data not available; NR: not reached; amp: amplification; EFS: event-free survival (months).
Considering the underlying complexity of the genomic landscape of TNBC, analysis of single mutations in a putative driver or known oncogenic pathway is likely insufficient (40). Different processes, such as age, exposure to carcinogens, DNA replication errors, defects in DNA repair, and the family of APOBEC cytidine deaminases, imprint patterns of mutations known as mutational signatures on the cancer genome. Whole-genome sequencing of 21 breast tumors initially showed the presence of five different mutational signatures in breast cancer, including focal hypermutation and APOBEC (40). More recently, the expanded analysis of 560 breast cancers revealed somatic base substitutions, indels, rearrangements, and copy number alterations in 93 candidate driver genes (41). Of the 10 most frequently mutated genes that accounted for 62% of drivers in the overall set, TP53, MYC, PTEN, ERBB2, and RB1 appeared enriched in the ER-negative cohort. Application of mathematical algorithms discriminated twelve base-substitution signatures (including the five previously identified signatures), two indel signatures and six rearrangements signatures. Large tandem duplications (>100 kb) were associated with rearrangement signature 1, mostly found in TP53-mutated, triple-negative tumors with high homologous recombination-deficiency (HRD) index but without BRCA1/2 mutations or BRCA1 promoter hypermethylation. In contrast, 91% of cases of with BRCA1 mutation or promoter hypermethylation fell into rearrangement signature 3, characterized predominantly by small tandem duplications (<10 kb). Additional research is required to fully understand the prognostic and therapeutic implications of these signatures.
Targeting genetically-altered signaling pathways in TNBC
Tumors with genetic alterations that promote activation of the PI3K pathway, found at a higher frequency in TNBC cell lines classified as LAR and mesenchymal-like, demonstrate in vitro and in vivo sensitivity to BEZ235 (a dual PI3K and mTOR inhibitor) (15). Loss of PTEN and INPP4B, which also sensitizes cell lines to PI3K inhibition (42), are more common in basal-like tumors (36). Oral pan-PI3K inhibitors, such as buparlisib (BKM120), or selective p110α-PI3K inhibitors, including alpelisib (BYL719) or taselisib (GDC-0032), have shown enhanced clinical activity in ER+ PIK3CA-mutant breast cancer, though fewer studies have been conducted in TNBC. In the BELLE-4 trial, patients with locally advanced or metastatic HER2− breast cancer were randomized to buparlisib or placebo in combination with paclitaxel as first-line therapy (43). Stratification was performed according to PI3K pathway activation, defined as PIK3CA mutation (detected by Sanger sequencing in exons 1, 7, 9, or 20) and/or low PTEN expression (1+ in ≤10% tumor cells). Approximately 25% of all enrolled patients (99/416) had hormone receptor-negative disease (i.e., TNBC), and of these, 36 (36.4%) had tumors considered to be PI3K-pathway activated. Addition of buparlisib to paclitaxel failed to demonstrate a significant improvement in progression-free survival (PFS) in the overall population or in those with PI3K-activated tumors. In patients with TNBC, there was a trend toward shorter median PFS with buparlisib compared to placebo (5.5 versus 9.3 months, respectively).
Ipatasertib, a highly selective AKT inhibitor, was evaluated in the phase II randomized trial LOTUS in combination with paclitaxel as first-line metastatic treatment for unselected TNBC (44). Ipatasertib improved PFS in the intent-to-treat population, and a similar trend was also noted in patients with PTEN-low tumors (IHC 0 in ≥50% tumor cells). In a prespecified analysis in patients with PIK3CA/AKT/PTEN-altered tumors (presence of activating PIK3CA/AKT1 mutations or PTEN-inactivating alterations using targeted NGS), median PFS with ipatasertib plus paclitaxel was 9.0 months versus 4.9 months in the placebo plus paclitaxel group, suggesting that the pathway may drive oncogenesis in a subset of patients with TNBC and providing the rationale for the ongoing randomized phase III IPATunity130 trial assessing the combination in pre-selected patients with activation of the PI3K pathway (NCT03337724). In addition, results from I-SPY 2, an adaptive-design trial testing novel agents in the neoadjuvant setting, showed an improvement in pCR with the addition of an allosteric AKT inhibitor, MK-2206, to standard chemotherapy in TNBC (40.2% versus 22.4% in control group), with a predicted 75.9% probability of success in a phase III trial (45).
Considering the higher prevalence of PI3K pathway aberrations in mesenchymal TNBC, of which 10–30% are metaplastic, a phase I study was conducted in this histologic subgroup to evaluate the combination of mTOR inhibition (temsirolimus or everolimus) with liposomal doxorubicin and bevacizumab (46). Responses were limited to patients with NGS aberrations in PIK3CA, AKT or PTEN. In the neoadjuvant setting, the addition of everolimus to cisplatin and paclitaxel did not increase pCR in molecularly unselected TNBC, and exploratory analyses showed that those who achieved pCR were not enriched for mutations in the PI3K/AKT/mTOR pathway (47).
Although alterations in genes encoding components of the RAS-MAPK pathway, such as KRAS, HRAS, BRAF, MEK1/2, are not observed as frequently in treatment-naïve TNBC as in other cancer types, EGFR is highly expressed in TNBC and can lead to upregulation of RAS-MAPK signaling (48). Across phase II and III trials, EGFR overexpression has not selected patients with TNBC who are more likely to derive benefit from EGFR-targeting monoclonal antibodies (e.g., cetuximab, panitumumab) or tyrosine kinase inhibitors (e.g., lapatinib) (49–52). Synergistic effects of combined RAF and MEK inhibition have been observed in MDA-MB-231 and MDA-MB-468 TNBC cell lines (53), likely due to the presence of an activating mutation in KRAS (codon 13) (54) and amplification of EGFR (55), respectively, in these cells. In addition, MYC (an oncogenic transcription factor that regulates transcriptional activity of multiple genes involved in cell proliferation, metabolism and survival) cooperates with RAS-MAPK to drive tumor progression in MCF10A triple-negative cell lines, and MEK inhibition potently inhibits tumor growth in MYC-overexpressed breast cancer (39). The presence of MYC amplification in 40% of basal-like tumors (36) suggests that MEK inhibition may be an attractive strategy in this selected population. Recently reported results from COLET, a randomized trial evaluating the MEK1/2 inhibitor cobimetinib with paclitaxel versus placebo and paclitaxel as first-line treatment for advanced TNBC showed a modest but not statistically significant increase in PFS (56). Selumetinib (MEK1/2 inhibitor) is also being tested in combination with vistusetib (mTORC1/2 inhibitor) in treatment-refractory solid tumors (NCT02583542). Although no objective responses were observed in the phase I trial, stable disease for >16 weeks was confirmed across tumor types, including TNBC (57).
As previously described, elevated expression of MYC has been identified across breast cancer types, with a strong association observed in triple-negative and basal-like tumors (58). Downregulation of MYC alone is insufficient to induce synthetic lethality and several combinatorial approaches have been investigated in preclinical models (59,60). Activation of the MYC pathway sensitizes TNBC cell lines to CDK inhibition, possibly by promoting cellular apoptosis through upregulation of BIM, a pro-apoptotic BCL-2 family member (58). CDK inhibitors, such as dinaciclib, downregulate MYC and a synergistic effect has been observed in combination with PARP inhibitors in MYC-driven TNBC cell lines, regardless of BRCA status (59). Other strategies focus on epigenetic modulation of gene transcription, such as inhibition and/or degradation of BET bromodomain proteins. BET inhibitors/degraders also induce downstream suppression of MYC and an apoptotic effect that is significantly enhanced when combined with small-molecule BCL-XL inhibitors (61,62). Altogether, these studies encourage further clinical research targeting MYC and exploring BET inhibitors in TNBC, and several clinical trials are ongoing in this area.
JAK-mediated activation of STAT transcription factors regulates transcriptional activity of targeted genes, including cell-cycle regulators (63), and the IL6/JAK2/STAT3 pathway plays an important role in the proliferation of CD44+CD24− stem-cell-like breast cancer cells, enriched in basal-like tumors (64). In TNBC cell lines, activation of JAK2/STAT5 has been implicated in PI3K/mTOR resistance and can be reversed by co-targeting both pathways (65). In addition, amplifications at the JAK2 locus (9p24) have been detected at a higher frequency in post-neoadjuvant TNBC samples compared to basal-like untreated tumors in TCGA, suggesting possible clonal selection after acquired chemotherapy resistance (39,66). Selective inhibition of JAK2 with NVP-BSK-805 (>20-fold selectivity of JAK2 over JAK1), administered with paclitaxel, significantly reduced pSTAT3 levels and tumor volume in vitro and in vivo compared to paclitaxel alone (66). In contrast, this effect was not observed with ruxolitinib (oral JAK1 and JAK2 inhibitor, with more limited activity against JAK2/STAT3) plus chemotherapy in JAK2-amplified TNBC cell lines. In a phase II trial in patients with metastatic TNBC, despite on-target inhibition and decreased pSTAT3 after two cycles of treatment, no responses were observed with single-agent ruxolitinib (67).
The NOTCH signaling pathway has been implicated in the differentiation and survival of stem cell-like tumor cells, and resistance to cytotoxic chemotherapy (68). Neutralizing antibodies targeting NOTCH1 significantly inhibit tumor growth in CD44+CD24− cells and enhance the activity of docetaxel (69). This synergistic effect with taxane-based therapy is also seen with PF-03084014, a reversible selective gamma-secretase inhibitor that blocks NOTCH signaling, in patient-derived TNBC xenograft models (70). Notch receptor mutations and focal amplifications are enriched in the triple-negative subtype, with most mutations either clustering in the heterodimerization domain or causing disruption of the PEST negative regulatory domain (71). These aberrations show evidence of pathway activation in TNBC and exhibit sensitivity to PF-03084014. In cell lines expressing NOTCH1 fusion alleles, gamma-secretase inhibition also downregulates expression of MYC and CCND1, two targets whose oncogenic role has been well-established in murine NOTCH-driven tumors (72). It is estimated that 13% of TNBC may be driven by these NOTCH-oncogenic alterations. In a phase Ib trial, 29 patients with molecularly-unselected treatment-refractory HER2-negative breast cancer (TNBC: n=26) were treated with PF-03084014 plus docetaxel. An objective response rate of 16% was confirmed among evaluable patients, and median PFS was 4.1 months in the expansion cohort (68).
As illustrated by the variable efficacy across clinical trials, the role that many of these genes play as potential oncogenic drivers in TNBC remains unclear. Many of these trials have not yielded clinically relevant improvements in outcomes. Although some of these studies show promising preliminary data for targeted therapies, many have yet to be explored either in larger, randomized studies or in populations enriched for molecular alterations. Also, up to 12% of TNBC carry low mutational burden and do not harbor mutations in known candidate driver or cytoskeletal genes (73), further highlighting the heterogeneity in the mutational landscape of TNBC and the need to improve our understanding of the functional implications of many of these alterations.
Germline BRCA-associated TNBC
Cancers that lack functional BRCA1 or BRCA2 have a deficiency in homologous recombination (HR) repair of DNA double-strand breaks (DSBs), leading to dependence on alternative mechanisms to repair these lesions, and genomic instability (74,75). Drugs that generate DSBs, such as alkylating agents (e.g., platinum, mytomicin C) or PARP inhibitors, cause persistent DNA damage in HR-deficient cells and, consequently, induction of cell cycle arrest and apoptosis (76,77). Germline mutations in BRCA1 or BRCA2 (BRCA1/2) are present in approximately 10% of patients with TNBC, and confer sensitivity to these drugs (78). In the previously mentioned TNT trial, despite failure to show a significant difference in activity between treatments in the overall population (n=376), in the 43 patients with deleterious BRCA1/2 germline mutations, carboplatin significantly improved ORR compared to docetaxel (68.0% vs. 33.3%, p=0.03) and PFS (6.8 vs. 4.4 months, interaction p=0.002) (12). In the neoadjuvant setting, elevated pCR rates (61–65%) have been observed with platinum agents in germline BRCA-associated TNBC, albeit BRCA-mutant patients in the GeparSixto trial obtained high pCR regardless of the addition of carboplatin (79,80).
Recently, PARP inhibitors (e.g., olaparib and talazoparib) have been compared to standard non-platinum chemotherapy in two phase III trials, OlympiAD and EMBRACA, respectively, in germline BRCA-associated metastatic HER2-negative breast cancer (81,82). Eligibility criteria included receipt of 2–3 previous lines of chemotherapy for metastatic disease, and receipt of an anthracycline and a taxane whether in the neoadjuvant, adjuvant or metastatic setting. Neoadjuvant or adjuvant platinum was allowed if the time that had elapsed since the last dose was 12 months in OlympiAD and 6 months in EMBRACA. Both trials enrolled a similar patient population, with some differences including the distribution of germline mutations (57.0% BRCA1 in OlympiAD; 54.5% BRCA2 in EMBRACA) and, concordantly, a slightly greater proportion of patients with hormone receptor-positive disease in EMBRACA (55.9%) than OlympiAD (50.3%). Results of both studies were positive, with improvements in ORR, PFS, and quality-of-life, favoring the PARP inhibitor. Compared to standard chemotherapy, a significant increase in median PFS was observed with olaparib (7.0 months versus 4.2 months, HR 0.58; p<0.001) and with talazoparib (8.6 months versus 5.6 months, HR 0.54; p<0.001). Safety profiles were also comparable across trials and hematological toxicity was the most common cause of dose modifications with PARP inhibition. An adjuvant trial (OlympiA, NCT02032823) in patients with germline BRCA-associated breast cancer is currently accruing. Of note, the reported response rates in the metastatic phase III trials of olaparib and talazoparib (59.9% and 62.6%, respectively) were similar to those previously reported with carboplatin, and platinum agents were not allowed in the chemotherapy control arm. At the present time, the comparative efficacy and optimal sequencing (given potentially overlapping resistance mechanisms) of PARP inhibitors versus platinum agents is unknown. In addition, whether PARP inhibitors may have activity in patients with other germline DNA repair defects (e.g., PALB2), or in patients with acquired somatic BRCA1/2 deleterious mutations, is unknown but is being tested in an ongoing clinical trial (NCT03344965).
Multiple mechanisms underlie the development of primary and acquired resistance to both platinum agents and PARP inhibitors, many of which have also been well-characterized in ovarian or prostate cancer. Molecular alterations leading to therapeutic resistance include, for example: small insertions/deletions that result in frameshift mutations and synthesis of truncated proteins (e.g., inherited founder mutation BRCA1185delAG) (83); secondary BRCA reversion mutations that reinstate HR-proficiency through restoration of the open reading frame and BRCA re-expression (84); exon 11 deletion splice variants that produce truncated, hypomorphic proteins (85); or point mutations in PARP1 that alter PARP trapping (86). In addition to genomic alterations, epigenetic changes such as loss of BRCA1 promoter hypermethylation via BRCA1 locus fusion rearrangements, with subsequent BRCA1 re-expression, have also been described after acquired resistance to DNA damaging drugs, including platinum or olaparib (87).
Several strategies to exploit potential synthetic lethality in HR-deficient tumors are being explored across solid tumors, including clinical trials combining PARP inhibitors with PI3K/AKT inhibitors (NCT02208375), immune checkpoint inhibition (NCT02657889) and HSP90 inhibitors (NCT02898207). HSP90 is a chaperone that assists in intracellular protein homeostasis by mediating protein folding and stabilization. HSP90 inhibitors block adequate protein folding, leaving the ¨client¨ protein (e.g., BRCA1) in the cytoplasm to be degraded by the proteasome. In vitro, HSP90 inhibition results in loss of BRCA1 expression and function and impaired DSB repair, sensitizing tumors to DNA damaging agents (88). Stabilizers of G-quadruplex DNAs such as CX-5641 bind to G4 DNA structures, interfering with progression of DNA replication complexes and inducing single-strand breaks that require HR for repair; thus, in BRCA-deficient tumors, failure to repair DNA damage leads to lethality, including in taxane-resistant BRCA1/2-deficient TNBC patient-derived xenograft models (89). Given its promising in vivo activity, CX-5461 is currently being explored in a phase I trial, with an expansion phase for unresectable breast cancer in patients with known BRCA1/2 or HRD germline aberrations (NCT02719977).
¨BRCAness¨ in sporadic TNBC
Somatic mutations and epigenetic alterations that inactivate BRCA1/2 and other DNA repair genes have been identified in sporadic cancers (90). Given that HR deficiency exposes specific therapeutic vulnerabilities, the detection of sporadic tumors with this so-called ¨BRCAness¨ phenotype could have clinical implications. Most BRCA1-related tumors are basal-like (91), and there is a marked resemblance in phenotype and biology between sporadic basal-like tumors and BRCA-associated cancers (90). Despite these similarities, targeting the HR pathway in sporadic basal-like cancer has revealed conflicting data in the metastatic and neoadjuvant settings. High-HRD score or basal phenotype (by PAM50 or IHC) did not predict greater benefit from carboplatin in TNT (12). Similarly, gene expression profiles were not associated with response to platinum in TBCRC-009, although a genomic instability signature based on HRD assays discriminated metastatic TNBC responders from non-responders (92). HR-deficiency (i.e. high-HRD score or tumor BRCA mutation) predicted increased pCR to neoadjuvant platinum (93–95). In GeparSixto, the addition of carboplatin to paclitaxel/liposomal doxorubicin improved pCR in HR-deficient tumors (64.9% vs. 45.2%, p=0.025), but not in HR-proficient tumors (40.7% vs. 20%, p=0.146) (94). Discrepancies across trials may be explained by significantly less methylated BRCA1/2 in metastases than in primary tumors, leading to potential loss of HR deficiency (96). Treatment exposure to alkylating agents commonly used in early-stage TNBC could drive clonal selection of HR-proficient cells less likely to respond to platinum in the metastatic setting. However, another explanation for these observed differences could be the robustness of the genomic metrics used to calculate HRD scores. With advances in sequencing technologies, an algorithm using whole-genome sequencing, also known as the HRDetect model, identified six mutational signatures present in germline BRCA1/2-mutated tumors that were then found to also predict HR-deficiency in sporadic tumors in the Sanger dataset (97). This aggregated BRCAness score was independently associated with benefit from platinum-based chemotherapy after adjusting for germline BRCA status and treatment timing, although the relatively small sample size (33 patients with metastatic breast cancer treated with either carboplatin or cisplatin as single-agent or in combination regimens) and the retrospective nature of the study (clouding the ability to establish a causal relationship) are limitations to be considered (98). Direct comparisons of these different measures should be further evaluated in ongoing prospective trials in HR-deficient breast cancer.
Currently, we lack predictive biomarkers to guide the choice of chemotherapy in sporadic basal-like TNBC, which comprises the majority of TNBC. Beyond germline BRCA mutations and the recent approval of olaparib and talazoparib in these patients, much remains unknown about the BRCAness features that may confer sensitivity to PARP inhibitors and DNA-damaging agents. Trials assessing these drugs are ongoing both in unselected and biomarker-selected populations (Table 3). In addition, preclinical data have demonstrated upregulation of PD-L1 expression after exposure to PARP inhibition in triple-negative MDA-MB-231 cells, with subsequent re-sensitization to a PARP inhibitor when combined with a PD-L1 antibody (99). Furthermore, the accumulation of cytosolic damaged DNA induced by PARP inhibition activates the STING pathway which, in turns, increases the expression of type-I IFN signaling and immune cell infiltration, regardless of BRCA mutational status (100). Altogether, this has provided the rationale to explore the combination of niraparib, a PARP inhibitor, and pembrolizumab, a PD-1 inhibitor, in the phase II clinical trial TOPACIO. Results from the TNBC cohort showed promising activity with an ORR of 28% in the 46 evaluable patients, and durable responses irrespective of tumor BRCA status, PD-L1 status or prior platinum exposure, although the highest ORR was observed in patients with tumor BRCA1 or BRCA2 mutations (60%) (101). A randomized phase II trial comparing olaparib in combination with the PD-L1 inhibitor atezolizumab versus olaparib alone in patients with BRCA-associated metastatic TNBC is currently ongoing (NCT02849496).
Table 3.
Ongoing clinical trials in BRCA-mutant or BRCAness-associated triple-negative breast cancer
|
Clinicaltrials.gov
Identifier |
Title | BRCA status eligibility criteria | Phase |
|---|---|---|---|
| Neoadjuvant | |||
| NCT03109080 | A Phase I of Olaparib With Radiation Therapy in Patients with Inflammatory, Loco-regionally Advanced or Metastatic TNBC or Patient with Operated TNBC With Residual Disease | BRCA mutation not required. | I |
| NCT03329937 | An Open-Label, Single-arm Pilot Study Evaluating the Antitumor Activity and Safety of Niraparib as Neoadjuvant Treatment in Localized, HER2-negative, BRCA-mutant Breast Cancer Patients | Deleterious or suspected deleterious BRCA1 or BRCA2 mutation (germline or somatic). | I |
| NCT02978495 | Neoadjuvant Carboplatin in Triple Negative Breast Cancer - A Prospective Phase II Study (NACATRINE Trial) | BRCA mutation not required. Includes BRCA-mutant specific cohorts. | II |
| NCT02789332 | A Randomized Phase II Trial to Assess the Efficacy of Paclitaxel and Olaparib in Comparison to Paclitaxel / Carboplatin Followed by Epirubicin/Cyclophosphamide as Neoadjuvant Chemotherapy in Patients with HER2-negative Early Breast Cancer and Homologous Recombination Deficiency | BRCA deleterious tumor or germline mutation and/or high HRD score. | II |
| NCT03150576 | Randomized, Phase II/III, 3 Stage Trial to Evaluate the Safety and Efficacy of the Addition of Olaparib to Platinum-based Neoadjuvant Chemotherapy in Breast Cancer Patients with TNBC and/or gBRCA. | TNBC or germline BRCA mutation HER2-negative breast cancer. | II/III |
| Adjuvant | |||
| NCT02032823 | A Randomized, Double-blind, Parallel Group, Placebo-controlled Multi-center Phase III Study to Assess the Efficacy and Safety of Olaparib Versus Placebo as Adjuvant Treatment in Patients With gBRCA1/2 Mutations and High Risk HER2 Negative Primary Breast Cancer Who Have Completed Definitive Local Treatment and Neoadjuvant or Adjuvant Chemotherapy | Suspected deleterious or deleterious BRCA1 and/or BRCA2 germline mutation. | III |
| Locally Advanced, Recurrent or Metastatic | |||
| NCT02950064 | Escalation Study of BTP-114 in Patients with Advanced Solid Tumors and BRCA or DNA Repair Mutation | Deleterious germline or somatic BRCA mutation or DNA-repair mutation. Abnormal HRD tests are also allowed. | I |
| NCT00576654 | A Phase I Dose-Escalation Study of Oral ABT-888 (NSC #737664) Plus Intravenous Irinotecan (CPT-11, NSC#616348) Administered in Patients with Advanced Solid Tumors | BRCA mutation not required. Includes BRCA-mutant specific cohort. | I |
| NCT02227082 | Olaparib Dose Escalation in Combination with High Dose Radiotherapy to the Breast and Regional Lymph Nodes | BRCA mutation not required. | I |
| NCT02898207 | A Phase 1 Study of PARP Inhibitor Olaparib and HSP90 Inhibitor AT13387 for Treatment of Advanced Solid Tumors with Expansion in Patients with Recurrent Epithelial Ovarian, Fallopian Tube, Peritoneal Cancer or Recurrent TNBC | BRCA mutation not required. Dose expansion excludes germline BRCA1 or BRCA2 mutations. | I |
| NCT03075462 | An Open, Non-randomized, Multi-center Phase I Study to Assess the Safety and Efficacy of Fluzoparib Given in Combination with Apatinib in Patients with Recurrent Ovarian Cancer or TNBC | BRCA mutation not required. | I |
| NCT03109080 | A Phase I of Olaparib With Radiation Therapy in Patients with Inflammatory, Loco-regionally Advanced or Metastatic TNBC or Patient with Operated TNBC With Residual Disease | BRCA mutation not required. | I |
| NCT03101280 | A Phase IB Combination Study of Rucaparib (CO-338) and Atezolizumab (MPDL3280A) in Participants with Advanced Gynecologic Cancers and TNBC | Part 1: All-comers; Part 2: deleterious germline or somatic BRCA mutation, or wild-type tumor BRCA but high levels of LOH | I |
| NCT02393794 | Phase I/II Study of Cisplatin Plus Romidepsin and Nivolumab in Metastatic Triple Negative Breast Cancer or BRCA Mutation-Associated Locally Recurrent or Metastatic Breast Cancer | TNBC or germline BRCA mutation breast cancer. | I/II |
| NCT02264678 | A Modular Phase I, Open-Label, Multicenter Study to Assess the Safety, Tolerability, Pharmacokinetics and Preliminary Anti-tumor Activity of AZD6738 in Combination with Cytotoxic Chemotherapy and/or DNA Damage Repair/Novel Anti-cancer Agents in Patients with Advanced Solid Malignancies | Cohort HER2-negative breast cancer: with BRCA mutation (germline or somatic); Cohort TNBC: without known BRCA mutation. | I/II |
| NCT02484404 | Phase I/II Study of the Anti-Programmed Death Ligand-1 Antibody MEDI4736 in Combination with Olaparib and/or Cediranib for Advanced Solid Tumors and Advanced or Recurrent Ovarian, Triple Negative Breast, Lung, Prostate and Colorectal Cancers | TNBC cohort requires germline BRCA1 or BRCA2 mutation. | I/II |
| NCT02401347 | A Phase II Clinical Trial of the PARP Inhibitor Talazoparib in BRCA1 and BRCA2 Wild-Type Patients With (i) Advanced Triple-Negative Breast Cancer and Homologous Recombination Deficiency (HRD), and (ii) Advanced HER2-Negative Breast Cancer or Other Solid Tumors with Either a Mutation in Homologous Recombination (HR) Pathway Genes | No deleterious BRCA mutation. TNBC with high HRD score or HER2-negative breast cancer with germline or somatic mutation in HR pathway. | II |
| NCT02203513 | A Phase II Single Arm Pilot Study of the Chk1/2 Inhibitor (LY2606368) In BRCA1/2 Mutation Associated Breast or Ovarian Cancer, TNBC, and High Grade Serous Ovarian Cancer | TNBC or germline BRCA mutation breast cancer. | II |
| NCT03205761 | A Phase II Clinical Trial to Analyze Olaparib Response in Patients with BRCA1 and/or 2 Promoter Methylation Diagnosed of Advanced Breast Cancer | Absence of deleterious or suspected deleterious germline BRCA mutations. Documented BRCA1 and/or BRCA2 promoter methylation. | II |
| NCT03330847 | A Phase II, Open Label, Randomized, Multi-center Study to Assess the Safety and Efficacy of Agents Targeting DNA Damage Repair in Combination with Olaparib Versus Olaparib Monotherapy in the Treatment of Metastatic TNBC Patients Stratified by Alterations in Homologous Recombinant Repair (HRR)-Related Genes (Including BRCA1/2) | BRCA mutation not required. Stratification by mutation in BRCA and HRR genes. | II |
| NCT02595905 | Phase II Randomized Placebo-Controlled Trial of Cisplatin with or Without ABT-888 (Veliparib) in Metastatic TNBC and/or BRCA Mutation-Associated Breast Cancer, With or Without Brain Metastases | TNBC or germline BRCA mutation breast cancer. | II |
| NCT01898117 | Biomarker Discovery Randomized Phase IIb Trial with Carboplatin-cyclophosphamide Versus Paclitaxel with or Without Atezolizumab as First-line Treatment in Advanced TNBC | BRCA mutation not required. | II |
| NCT03414684 | A Randomized Phase II Trial of Carboplatin with or Without Nivolumab in First- or Second-line Metastatic TNBC | BRCA mutation not required. Stratification by germline BRCA mutation. | II |
| NCT02498613 | A Phase 2 Study of Cediranib in Combination with Olaparib in Advanced Solid Tumors | BRCA mutation not required. | II |
Clinicaltrials.gov database was searched for interventional-only clinical trials that are recruiting as of April 14, 2018. Only drug-based interventions were considered. Search terms included ¨triple negative breast cancer¨, ¨HER2-negative breast cancer¨, ¨BRCA¨, and ¨PARP¨.
Epigenetic markers and therapies in TNBC
Epigenetic alterations, including changes in DNA methylation of gene promoter regions and post-translational modification of histone proteins, are a recognized hallmark of cancer. Approximately 60–80% of basal-like and claudin-low breast cancers have aberrant DNA hypermethylation (102). Compared to luminal and HER2-positive cancers, TNBC exhibits extensive CpG methylation of the promoter regions of nine epigenetic biomarker genes (CDH1, CEACAM6, CST6, GNA11, ESR1, MUC1, MYB, SCNN1A, and TFF3). DNA hypermethylation-dependent silencing of these genes is associated with worse RFS across all molecular subtypes and stages, compared to breast cancers unmethylated for these genes (40% RFS at 70 and 30 months, respectively). A non-significant trend toward RFS disadvantage has also been described among basal-like and claudin-low tumors that have this 9-gene methylation signature (102). In addition, promoter hypomethylation of three breast cancer stem cell-related genes, (CD44, CD133, and MSH1), which strongly correlates with positive IHC staining and thus gene activation, has been shown to predict triple-negative status (103). Differences in histone modifications are also associated with differences in the expression of breast cancer genes across subtypes, separating luminal tumors, enriched with H3K27me3-modified genes, from non-luminal tumors (TNBC/HER2-positive), enriched with H3K9ac-regulated genes (104).
Therapies targeting epigenetic modifications, such as inhibitors of DNA methyltransferases (DNMT; 5-azacitidine, decitabine) and histone deacetylases (HDAC; entinostat, vorinostat), have yielded disappointing results to date in TNBC. The combination of 5-azacitidine and entinostat did not achieve any responses among 13 women with advanced TNBC treated in a phase II study (105). No significant changes in gene expression in paired biopsies before and after two months of treatment were observed, possibly due to absent ER promoter DNA methylation at baseline. Novel approaches in epigenetic modulation include BET bromodomain inhibitors that bind to acetylated lysine residues in histones, displacing bromodomain proteins from chromatin and inhibiting transcriptional activity (106). BET inhibitors achieve potent suppression of tumor growth in TNBC cell lines characterized by more basal-like and claudin-low/stem cell-like features (61). Several BET inhibitors are currently in early stages of clinical testing as single-agents or in combination with immunotherapy (NCT01587703, NCT02391480, NCT02711137).
Immune subtypes of TNBC
Increasing data suggest that the immune system is critical for disease outcome in TNBC. Analyses from neoadjuvant and adjuvant TNBC trials have shown that tumor-infiltrating lymphocytes (TIL), assessed by hematoxylin-eosin staining, are predictive of response to therapy and strongly associated with improved survival (107,108). Stratification of TNBC based on quantitative TIL evaluation has distinguished immune ¨hot¨ (high-TIL) and ¨cold¨ (low-TIL) tumors, which also appear to correlate with response to immune-checkpoint inhibitors in the metastatic setting (109). Paired biopsies pre- and post-neoadjuvant therapy have shown that the immune microenvironment can be modulated by chemotherapy, converting tumors from ¨cold¨ to ¨hot¨, and these cases with highly-infiltrated residual TNBC have improved survival (110). Phenotypic TIL characterization has also provided further insight into the populations of immune cells (e.g. CD8+ T-cells; elevated CD8/FOXP3 ratio) that may be responsible for this positive effect (111). Elevated expression in TNBC of immune markers of tumor evasion PD-1/PD-L1 has prompted clinical assessment of inhibitors of these checkpoints, with modest efficacy as monotherapy and encouraging results in combination with chemotherapy (Table 4) (109,112–118).
Table 4.
Results of PD-1/PD-L1 Inhibition in Advanced Triple-Negative Breast Cancer
| SINGLE-AGENT IMMUNOTHERAPY | COMBINATION WITH CHEMOTHERAPY | ||||||
|---|---|---|---|---|---|---|---|
| ANTI-PD-1 | ANTI-PD-L1 | ANTI-PD-1 | ANTI-PD-L1 | ||||
| Pembrolizumab in PD-L1+ TNBC (KEYNOTE-012) (111) |
Pembrolizumab in Metastatic TNBC (KEYNOTE-086) (112,113) |
Atezolizumab in TNBC Unselected for PD-L1 (108) |
Avelumab in TNBC Unselected for PD-L1 (JAVELIN) (116) |
Eribulin +/− Pembrolizumab in Metastatic TNBC (ENHANCE-1/ KEYNOTE-150) (115) |
Atezolizumab + nab-Paclitaxel in TNBC Unselected for PD-L1 (114) |
Atezolizumab + nab- Paclitaxel vs. Placebo + nab-Paclitaxel in Metastatic TNBC (Impassion130) (117) |
|
| TUMOR CHARACTERISTICS | |||||||
| Definition of PD-L1 positivity | ≥1% TC or any staining in stroma | ≥1% TC or any staining in stroma | ≥5% IC | ≥1% TC; ≥10% IC | ≥1% TC or any staining in stroma | ≥1% TC; ≥1% IC | ≥1% IC |
| PD-L1 status inclusion criteria | Positive | All-comers Cohort A: pre-treated, any PD-L1 Cohort B: untreated, PD-L1+ |
All-comers | All-comers | All-comers Stratum 1: no prior therapy Stratum 2: 1-2 prior lines |
All-comers | All-comers |
| Frequency of PD-L1 positivity among evaluable cases (%) | 65/111 (58.6) | A: 105/169 (62.1) B: 128/207 (61.8) |
71/108 (65.7) | TC: 33/48 (68.8) IC: 9/48 (18.8) | 49/98 (50.0) | IC: 11/21 (52.4) | 369/902 (40.9) |
| PATIENT CHARACTERISTICS | |||||||
| Total number of patients enrolled | 32 | A: 170 B: 84 |
115 | 58 | 107 (S1: 66; S2: 41) | 32 | 902 |
| Total number of patients included in efficacy analysis | 27 | A: 170 B: 84 |
112 ‡ | 58 | 107 (106 ‡) | 32 | 902 |
| Median prior lines of therapy in metastatic setting (range) | 2 (0-9) | A: NA B: 0 |
7 (0-21) | NA* | S1: 0 S2: 1-2 |
5 (1-10) | 0 |
| EFFICACY | |||||||
| ORR, % | 18.5 | A: 4.7 B: 22.6 |
Overall: 9.8 1st line: 26.3 2nd line: 3.6 3rd/+ line: 7.7 |
5.2 | Overall: 26.4 S1: 29.2 S2: 22.0 |
Overall: 37.5 1st line: 46.1 2nd line: 22.2 3rd/+line: 40.0 |
ITT: 56.0 vs. 45.9 |
| ORR in PD-L1+ cohort, % | 18.5 | A: 4.8 B: 22.6 |
12.7 | 22.2** | 30.6 | 36.3** | 58.9 vs. 42.6 |
| CBR, % | 25.9 † | A: 7.6 B: 25.0 |
NA | 31.0 † | Overall: 36.8 S1: 40.0 S2: 31.7 |
81.3 † | NA |
| Median PFS, mo. (95% CI) |
1.9 (1.7-5.5) | A: 2.0 (1.9-2.0) B: 2.1 (2.0-2.3) |
NA | 1.5 (1.4-1.7) | Overall: 4.2 (4.1-5.6) S1: 4.9 (4.1-6.1) S2: 4.1 (2.1-6.2) |
NE | ITT: 7.2 vs. 5.5; HR 0.80 (0-69-0.92) PD-L1+: 7.5 vs. 5.0; HR 0.62 (0.49-0.78) |
| Median OS, mo. (95% CI) |
11.2 (5.3-NR) | A: 8.9 (7.2-11.2) (CR, PR, or SD: NR; PD: 7.1 [6.3-8.8]) B: 19.2 (11.3-NE) |
9.3 (7.0-12.6) | 9.2 (4.3-NE) | Overall: 17.7 (13.7-NE) S1: 17.7 (13.3-NE) S2: 16.3 (12.4-19.2) |
NE (8.0-NE) |
ITT: 21.3 vs. 17.6; HR 0.84 (0.69-1.02) PD-L1+: 25.0 vs. 15.5; HR 0.62 (0.45-0.86) |
TNBC: triple-negative breast cancer; TC: tumor cells; IC: immune cells; ORR: objective response rate; CBR: clinical benefit rate (defined as complete response, partial response or stable disease for ≥ 24 weeks); mo.: months; NR: not reached; NE: not estimable; NA: not available; PFS: progression-free survival; OS: overall survival; CI: confidence interval; ITT: intent-to-treat population; HR: hazard ration.
Response rates per RECIST 1.1 criteria.
DCR: defined as confirmed complete, partial response or stable disease as best response.
Number of patients considered Objective Response-Evaluable.
In overall population, the median number of prior lines of therapy in any setting was 4 (range 1-10). In TNBC cohort, 50% had received ≥2 prior lines of therapy for metastatic disease.
According to PD-L1 positivity in IC.
Recently, results from a large phase III trial (IMpassion130) that randomized patients in the first-line TNBC metastatic setting to receive nab-paclitaxel combined with either atezolizumab (PD-L1 inhibitor) or placebo were reported (118). While the absolute difference in median PFS in the PD-L1-positive population (2.5 months) was not strikingly different than that seen in the intent-to-treat (ITT) cohort (1.7 months), at a median follow up of 12.9 months, a 9.5-month clinically meaningful improvement in median OS was noted in patients with PD-L1-positive tumors, in contrast to a 3.7-month difference in the ITT population (118). No PFS or OS differences were noted in the subset of patients with PD-L1-negative tumors (119). Several other randomized trials have completed accrual and are awaiting data maturity to report. Whether similar results may be achieved with chemotherapy plus immunotherapy in later lines is unknown at this time. Of note, increased ORR have been observed in patients with previously untreated metastatic TNBC with monotherapy PD-1/PD-L1 inhibitors, suggesting that these agents may be more active in less heavily pre-treated metastatic disease (120).
Efforts to identify patients with tumors that are more or less likely to benefit from immunotherapy-based approaches are ongoing. As evidenced in the IMpassion130 trial, not all patients with PD-L1 tumors (defined by the presence of ≥1% IHC staining on immune cells) respond to PD-L1 inhibition and, contrarily, there are patients who despite negative PD-L1 staining, appear to derive benefit from treatment. Beyond immunohistochemical classifications, genetic alterations of immune-regulatory genes have also segregated TNBC into subgroups with different prognostic and possibly therapeutic implications. CD274 (encoding PD-L1) and PDCD1LG2 (encoding PD-L2) genes localize to the 9p24 locus, adjacent to JAK2, constituting the PDJ amplicon. Overexpression of PD-L1 is observed in 88% of tumors with amplifications in the 9p24/JAK2 locus, which are found at higher frequency in post-neoadjuvant residual TNBC (66). In TNBC, the PDJ amplicon identified a subset of patients at significantly greater risk of recurrence (121), and could be a potential biomarker for selection of high-risk patients who may benefit from PD-1/PD-L1 blockade. Activating mutations in the RAS/MAPK pathway, present in 15% of residual disease, correlated with reduced TIL; inhibition of MEK upregulated PD-L1 expression, synergizing with PD-1/PD-L1 antibodies in murine models (122). Furthermore, high tumor mutational burden has been associated with improved outcomes with PD-1 inhibition in other cancer types (123), and may represent an independent biomarker of response.
Transcriptomic analysis of tumor-associated stroma in TNBC has revealed the presence of four axes, each with differential expression of genes related to T-cell, B-cell, epithelial (E) and desmoplasia (D) markers. The E-axis inversely correlated with LAR Lehmann-subtype, and the D-axis was positively associated with MSL while also determining the prognostic value of T-, B-, and E-axes (124). Furthermore, these axes strongly influenced the location of CD8+ TIL (125), which may impact antitumoral response to immune-checkpoint inhibitors. Similarly, when analyzing the tumor compartment, the presence of the immunomodulatory signature (associated with elevated lymphocytic infiltration and increased expression of immune checkpoint regulators, e.g. PD-1/PD-L1) (18), significantly differs across refined TNBCtypes, with the highest rates observed in BL1 (48%) and the lowest in M (0%) (126). Whether transcriptomic profiling could be incorporated to routine clinical practice to help select TNBC patients with a greater likelihood of responding to immune checkpoint inhibitors, similar to the applicability of gene expression assays (e.g., 21-gene Recurrence Score, Oncotype®) to predict chemotherapy benefit in ER-positive breast cancer (127), remains to be seen.
To date, one single marker has not been proven to effectively select patients who are more likely to respond to immunotherapies. Recently, the development of multiplexed imaging techniques has enabled analysis of the spatial distribution and interaction between tumor and immune cells, showing that in TNBC there is high intratumor topologic heterogeneity for the expression of PD-1 on cytotoxic CD8+ and helper CD4+ T-cells (128). Tumors with immune cells that are spatially separated from tumor cells, also defined as compartmentalized (as opposed to mixed immune cells with tumor cells), predominantly express PD-1 on CD4+ T-cells and are independently associated with improved survival. Given the complexity of these interactions, integration of comprehensive omics analyses of samples with detailed clinical data annotation will be needed to better understand how the relationship between the tumor and its microenvironment impacts response to treatment.
Evolutionary paths of TNBC
Analyses of paired primary and metastatic TNBC samples are also needed to better understand the drivers of disease progression. Clonal frequencies vary significantly across TNBC at the time of diagnosis, suggesting their occurrence at different stages of tumorigenesis (73). There is limited sequencing data in metastatic triple-negative tumors and much remains unknown about the differences in the molecular landscape of TNBC over its natural history. Multiclonal seeding from different cell populations in the primary to the metastasis has been reported in two cases of basal-like TNBC, where, in addition, most putative driver mutations were shared, rather than acquired, between primary and metastatic lesions (129,130). Also, most TNBC primary tumors and metastases are polyclonal, with overlapping clones, suggesting that polyclonal metastasis is common in TNBC. Phylogenetic analysis has the potential to distinguish local recurrences from second primary tumors and to help determine the origin of a metastatic lesion in a patient with history of independent primary tumors (131). Given the differences in management of primary and recurrent tumors, sequencing of longitudinal samples could impact treatment decisions.
Receptor status, according to IHC, and also intrinsic subtype, can change at time of recurrence (132), but the clinical relevance of molecular phenotype switch remains unclear and IHC-subtypes largely drive current treatment decisions in breast cancer. Loss of ER and PR expression occurs in approximately 10–12% of asynchronous recurrences, inducing a switch to TNBC in the metastasis (133), and has been associated with worse survival compared to cases with concordant hormone receptor-positive recurrence (134). To date, we do not fully understand the mechanisms that cause this conversion, and if there are special considerations that should be made when treating this patient population. Of note, there are also breast tumors that express low levels (1–9%) of ER and PR, and it remains unclear whether these cases derive significant benefit from endocrine therapy (135). Retrospective studies have shown that almost half of tumors with 1–9% ER staining are basal-like (136), suggesting that we should consider these tumors similar to TNBC and apply treatment algorithms, including enrollment onto clinical trials, for TNBC in these patients.
The extent of residual disease post-neoadjuvant chemotherapy, quantified per residual cancer burden index, is a well-established risk factor for recurrence (137). Residual disease has been used as a marker to select patients for escalation of adjuvant therapy, particularly in TNBC, based on the significant absolute improvement observed in patients treated with versus without capecitabine in terms of 3-year DFS (69.8% versus 56.1%, respectively; HR 0.58; 95% CI: 0.39–0.87) and OS (78.8% versus 70.3%, respectively; HR 0.52; 95% CI: 0.30–0.90) (138). However, not all patients with residual disease will recur. Distinguishing between the molecular mechanisms of chemoresistance and those that drive the development of metastatic disease remains a challenge. Intratumor genetic heterogeneity has been widely described in TNBC and may be associated with a decreased likelihood of achieving a pCR (139,140). Bulk exome and single-cell sequencing in a small number of pre- and post-neoadjuvant therapy samples suggest the occurrence of adaptive clonal extinction or persistence and acquired transcriptional reprogramming as potential models of chemoresistance (140). Other single-cell resolution studies support the hypothesis that most mutation and copy number events occur in early stages of tumor evolution, rather than develop gradually over time implying punctuated evolution (141). Validation of these findings in larger sets of tumors with associated long-term outcome data is key to understand the impact of genomic and phenotypic evolution of triple-negative cancer cells.
Conclusions
In summary, TNBC is comprised of a broad spectrum of biologically distinct subtypes with overlapping alterations. Despite advances in tumor characterization, separately, each classification has not yet translated to specific treatments or choices of treatments, with the exception of PARP inhibitors or platinum agents in germline BRCA1/2 carriers, and potentially in the near future immune checkpoint inhibition in tumors with PD-L1-positive immune cells. Comprehensive integrated analysis of data generated from different ¨omics¨ technologies may provide more insight into the etiology, evolution of TNBC and, possibly prevention and new treatment strategies. Nonetheless, as the volume of information exponentially increases, identifying alterations that are critical for tumor growth and survival continues to be a challenge. In addition, the utility of these profiles is largely limited by genetic and epigenetic heterogeneity within the tumor. There have been several large-scale efforts to find new targets, including shRNA/CRISPR screens (64,142–144). Using loss-of-function RNAi-based screens across over 500 cancer cell lines, biocomputational algorithms have been developed to help predict cancer dependencies (143), and novel potently selective inhibitors, as single-agents or in combination, will be needed to effectively block these targets (61,145,146). Similarly to cell lines and organoids, patient-derived xenografts enable high-throughput drug screening, but with the potential advantages of analyzing tumor growth metrics and characterizing drug response in models that retain the histopathologic features and inter- and intratumor genomic heterogeneity of the explanted tumor (147). Given the complexity of these techniques and sample size of individual cohorts, institutional collaborations should be forged to create biobanks that will provide a platform to help answer questions of interest in specific subsets of patients with TNBC.
Most trials to date have been performed in unselected TNBC, hoping to find a signal of efficacy in subgroup analyses. Prospective validation of biomarker-driven approaches has been widely considered a necessary step for approval of targeted therapies over the past years. Only recently were results published from the first trial in TNBC to prospectively stratify patients by the presence of a tumor gene signature (148). In this neoadjuvant study, patients were randomized to receive paclitaxel with or without LCL161, a small molecule antagonist of inhibitor of apoptosis proteins. LCL161 induces tumor necrosis factor (TNF)-mediated apoptosis, and preclinical work identified a 3-gene signature (elevated TNFα, elevated RIPK1 and reduced STK39) that was associated with sensitivity to LCL161. In patients with signature-positive tumors, the pCR rate was higher in the combination versus the control arm (38.2% vs. 17.2%, respectively), as opposed to lower pCR in those that were negative for the signature (5.6% vs. 16.4%, respectively), albeit with significant toxicity that led to treatment discontinuation in almost one-fifth of patients treated with LCL161 and paclitaxel (148). Of the total of 312 patients who signed consent for molecular prescreening for this trial, 207 had a valid signature score and were treated on study (of which 63 [30.4%] were found to be positive for the signature). Enrollment was completed in approximately 25 months but required participation of 47 international sites across 11 countries. Inability to ship samples for testing (4.2%) and assay failure (7.1%) were among the reasons for exclusion of patients, highlighting the challenges of prospectively implementing molecular testing in clinical trials, including those evaluating biomarkers with a prevalence as high as the 30% rate observed in this trial.
Another limitation of conducting single oncogene-driven clinical trials is the fact that there are complex interactions and overlap between different genomic alterations (e.g., comparable prognosis between PI3K-activated, TP53 wild-type TNBC and ER-positive breast cancer) with consequences that are not clearly understood to date nor taken into consideration in study designs. As the field of genomics in TNBC evolves and new insights are gained, these factors may need to be incorporated into trial designs, particularly when posthoc stratification by various forms of analysis may be needed to interpret and demonstrate subgroup effects.
As NGS, immune-profiling, and other technologies become widely available, biomarker-selected basket trials across multiple cancer types are of particular importance to evaluate the efficacy of matched targeted therapies. Considering the multiple molecular hypotheses for treatment, dynamic biomarker-adjusted platforms, such as WSG-ADAPT or I-SPY, aim to improve the efficiency of early drug development by predicting the probability of success in phase III clinical trials (45,149). However, given smaller and smaller potential subsets of interest, the success of biomarker-enriched designs will increasingly depend on more effective strategies to ensure that a larger pool of potentially eligible patients have the opportunity to be offered participation in such trials. Furthermore, given the evident heterogeneity in the molecular landscape of TNBC and current efforts to integrate omics data to better understand the underlying biological processes, the combinations of features in the tumor and its microenvironment that may be identified, and potentially targeted, seem endless. Conduction of randomized studies that require a large number of patients, aiming to test each individual hypothesis and demonstrate superiority of novel drugs to current standard-of-care regimens, is simply not feasible. As subsets of patients with rare, potentially actionable targets are identified, exploring multiple treatment options in these select populations is becoming more challenging, and this will likely translate into an increasing need to mindfully extrapolate results from subgroup analyses. Optimization of trial designs, including umbrella trials in TNBC (in which patients are assigned to an intervention based on genomic and/or immune profiling of the tumor at baseline), ¨pick-the-winner¨ strategies (with smaller sample sizes) and incorporation of comprehensive fresh biospecimen collection for correlative substudies, may provide proof-of-concept to help select therapies that are more likely to succeed in larger trials.
To overcome the challenges of limited, single-institution studies, multiple genomic data sharing initiatives such as Project GENIE (American Association of Cancer Research), Genomic Data Commons (National Cancer Institute) or cBioPortal have been developed as large repositories of sequencing data. Despite these efforts, one of the major limitations of these large-scale studies is the lack of detailed clinical annotation, making it difficult to answer specific questions such as the association between genomic features and prior exposure to therapy, changes in receptor subtype over time (i.e., due to absent ER, PR and HER2 status at different time points) or clinical outcomes (e.g., response, survival). Another limitation is the heterogeneity in the utilization of exome/genome versus targeted panel sequencing across cancer centers, which limits the ability to perform in-depth analyses to genes that are common to all panels. In upcoming years, we anticipate standardization of clinical sequencing across institutions and implementation of machine-learning tools that will help extract clinical data from electronic medical records, facilitating a seamless integration of genomic and clinical information in both private and public datasets.
Furthermore, to address the need for advances in drug development and biomarker discovery in TNBC, the elaboration of prospective, large-scale, longitudinal multi-center cohort studies in TNBC that have the ability to capture a patient´s clinical course and collect fresh-frozen tissue, blood and other biospecimens over a longer timeframe, over multiple treatments, regardless of trial participation, and across a larger number of patients, has the potential to vastly improve our knowledge of the dynamic changes in tumor biology, and the markers of response or resistance to treatment. These platforms may also be utilized to effectively communicate, offer and expand clinical trial participation to patients across collaborating institutions in order to help answer clinically relevant questions in a timely manner and, ultimately, improve outcomes in patients diagnosed with TNBC.
Significance.
Triple-negative breast cancer is characterized by higher rates of relapse, greater metastatic potential and shorter overall survival compared to other major breast cancer subtypes. The identification of biomarkers that can help guide treatment decisions in triple-negative breast cancer remains a clinically unmet need. Understanding the mechanisms that drive resistance is key to the design of novel therapeutic strategies to help prevent the development of metastatic disease and, ultimately, to improve survival in this patient population.
Acknowledgments
Grant Support: This work was supported in part by the National Cancer Institute R35CA197623 (K.P.).
Footnotes
Disclosure of Potential Conflicts of Interest: K.P. is a Scientific Advisory Board member of Mitra Biotech. N.U.L. receives research funding from Pfizer, Genentech, Kadmon, Novartis, Array Biopharma, and she is also a consultant to Genentech, Novartis, Seattle Genetics, and Daichii. No potential conflict of interest was disclosed by A.C.G.C.
REFERENCES
- 1.Bertucci F, Houlgatte R, Benziane A, Granjeaud S, Adelaide J, Tagett R, et al. Gene expression profiling of primary breast carcinomas using arrays of candidate genes. Hum Mol Genet 2000;9:2981–91. [DOI] [PubMed] [Google Scholar]
- 2.Brown M, Tsodikov A, Bauer KR, Parise CA, Caggiano V. The role of human epidermal growth factor receptor 2 in the survival of women with estrogen and progesterone receptor-negative, invasive breast cancer: the California Cancer Registry, 1999–2004. Cancer 2008;112:737–47. [DOI] [PubMed] [Google Scholar]
- 3.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007;13:4429–34. [DOI] [PubMed] [Google Scholar]
- 4.Andre F, Zielinski CC. Optimal strategies for the treatment of metastatic triple-negative breast cancer with currently approved agents. Ann Oncol 2012;23 Suppl 6:vi46–51. [DOI] [PubMed] [Google Scholar]
- 5.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 6.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010;12:R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2010;15 Suppl 5:39–48. [DOI] [PubMed] [Google Scholar]
- 8.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Molecular oncology 2011;5:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Res Treat 2012;135:301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Lluch A, Albanell J, Barry WT, Fan C, Chacon JI, et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br J Cancer 2014;111:1532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sikov WM, Barry WT, Hoadley KA, Pitcher BN, Singh B, Tolaney SM, et al. Abstract S4–05: Impact of intrinsic subtype by PAM50 and other gene signatures on pathologic complete response (pCR) rates in triple-negative breast cancer (TNBC) after neoadjuvant chemotherapy (NACT) +/− carboplatin (Cb) or bevacizumab (Bev): CALGB 40603/150709 (Allianc. Cancer Research 2015;75:S4–05-S4-. [Google Scholar]
- 12.Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 2018;24:628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruna A, Greenwood W, Le Quesne J, Teschendorff A, Miranda-Saavedra D, Rueda OM, et al. TGFbeta induces the formation of tumour-initiating cells in claudinlow breast cancer. Nat Commun 2012;3:1055. [DOI] [PubMed] [Google Scholar]
- 14.Bhola NE, Balko JM, Dugger TC, Kuba MG, Sanchez V, Sanders M, et al. TGF-beta inhibition enhances chemotherapy action against triple-negative breast cancer. J Clin Invest 2013;123:1348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bareche Y, Venet D, Ignatiadis M, Aftimos P, Piccart M, Rothe F, et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol 2018;29:895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masuda H, Baggerly KA, Wang Y, Zhang Y, Gonzalez-Angulo AM, Meric-Bernstam F, et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin Cancer Res 2013;19:5533–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 2016;11:e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echavarria I, Lopez-Tarruella S, Picornell A, Garcia-Saenz JA, Jerez Y, Hoadley K, et al. Pathological Response in a Triple-Negative Breast Cancer Cohort Treated with Neoadjuvant Carboplatin and Docetaxel According to Lehmann’s Refined Classification. Clin Cancer Res 2018;24:1845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaayvaz M, Cristea S, Gillespie SM, Patel AP, Mylvaganam R, Luo CC, et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun 2018;9:3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burstein MD, Tsimelzon A, Poage GM, Covington KR, Contreras A, Fuqua SA, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses’ Health Study. Mod Pathol 2011;24:924–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asghar US, Barr AR, Cutts R, Beaney M, Babina I, Sampath D, et al. Single-Cell Dynamics Determines Response to CDK4/6 Inhibition in Triple-Negative Breast Cancer. Clin Cancer Res 2017;23:5561–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res 2013;19:5505–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traina TA, Miller K, Yardley DA, Eakle J, Schwartzberg LS, O’Shaughnessy J, et al. Enzalutamide for the Treatment of Androgen Receptor-Expressing Triple-Negative Breast Cancer. J Clin Oncol 2018;36:884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnefoi H, Grellety T, Tredan O, Saghatchian M, Dalenc F, Mailliez A, et al. A phase II trial of abiraterone acetate plus prednisone in patients with triple-negative androgen receptor positive locally advanced or metastatic breast cancer (UCBG 12–1). Ann Oncol 2016;27:812–8. [DOI] [PubMed] [Google Scholar]
- 29.Traina TA, Yardley DA, Schwartzberg LS, O’Shaughnessy J, Cortes J, Awada A, et al. Overall survival (OS) in patients (Pts) with diagnostic positive (Dx+) breast cancer: Subgroup analysis from a phase 2 study of enzalutamide (ENZA), an androgen receptor (AR) inhibitor, in AR+ triple-negative breast cancer (TNBC) treated with 0–1 prior lines of therapy. Journal of Clinical Oncology 2017;35:1089-. [Google Scholar]
- 30.Lehmann BD, Bauer JA, Schafer JM, Pendleton CS, Tang L, Johnson KC, et al. PIK3CA mutations in androgen receptor-positive triple negative breast cancer confer sensitivity to the combination of PI3K and androgen receptor inhibitors. Breast Cancer Res 2014;16:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millis SZ, Gatalica Z, Winkler J, Vranic S, Kimbrough J, Reddy S, et al. Predictive Biomarker Profiling of > 6000 Breast Cancer Patients Shows Heterogeneity in TNBC, With Treatment Implications. Clin Breast Cancer 2015;15:473–81 e3. [DOI] [PubMed] [Google Scholar]
- 32.Bardia A, Mayer IA, Diamond JR, Moroose RL, Isakoff SJ, Starodub AN, et al. Efficacy and Safety of Anti-Trop-2 Antibody Drug Conjugate Sacituzumab Govitecan (IMMU-132) in Heavily Pretreated Patients With Metastatic Triple-Negative Breast Cancer. J Clin Oncol 2017;35:2141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modi S, Pusztai L, Forero A, Mita M, Miller K, Weise A, et al. Abstract PD3–14: Phase 1 study of the antibody-drug conjugate SGN-LIV1A in patients with heavily pretreated triple-negative metastatic breast cancer. Cancer Research 2018;78:PD3–14-PD3-. [Google Scholar]
- 34.Yardley DA, Weaver R, Melisko ME, Saleh MN, Arena FP, Forero A, et al. EMERGE: A Randomized Phase II Study of the Antibody-Drug Conjugate Glembatumumab Vedotin in Advanced Glycoprotein NMB-Expressing Breast Cancer. J Clin Oncol 2015;33:1609–19. [DOI] [PubMed] [Google Scholar]
- 35.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. Cancer genome landscapes. Science 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weisman PS, Ng CK, Brogi E, Eisenberg RE, Won HH, Piscuoglio S, et al. Genetic alterations of triple negative breast cancer by targeted next-generation sequencing and correlation with tumor morphology. Mod Pathol 2016;29:476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 2014;4:232–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fedele CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, Millar EK, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci U S A 2010;107:22231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin M, Chan A, Dirix L, O’Shaughnessy J, Hegg R, Manikhas A, et al. A randomized adaptive phase II/III study of buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel for the treatment of HER2- advanced breast cancer (BELLE-4). Ann Oncol 2017;28:313–20. [DOI] [PubMed] [Google Scholar]
- 44.Kim SB, Dent R, Im SA, Espie M, Blau S, Tan AR, et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2017;18:1360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tripathy D, Chien AJ, Hylton N, Buxton MB, Ewing CA, Wallace AM, et al. Adaptively randomized trial of neoadjuvant chemotherapy with or without the Akt inhibitor MK-2206: Graduation results from the I-SPY 2 Trial. Journal of Clinical Oncology 2015;33:524.25584001 [Google Scholar]
- 46.Basho RK, Gilcrease M, Murthy RK, Helgason T, Karp DD, Meric-Bernstam F, et al. Targeting the PI3K/AKT/mTOR Pathway for the Treatment of Mesenchymal Triple-Negative Breast Cancer: Evidence From a Phase 1 Trial of mTOR Inhibition in Combination With Liposomal Doxorubicin and Bevacizumab. JAMA Oncol 2017;3:509–15. [DOI] [PubMed] [Google Scholar]
- 47.Jovanovic B, Mayer IA, Mayer EL, Abramson VG, Bardia A, Sanders ME, et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients with Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-term Outcome Correlated with Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clin Cancer Res 2017;23:4035–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 2012;149:307–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yardley DA, Ward PJ, Daniel BR, Eakle JF, Lamar RE, Lane CM, et al. Panitumumab, Gemcitabine, and Carboplatin as Treatment for Women With Metastatic Triple-Negative Breast Cancer: A Sarah Cannon Research Institute Phase II Trial. Clin Breast Cancer 2016;16:349–55. [DOI] [PubMed] [Google Scholar]
- 50.Nabholtz JM, Chalabi N, Radosevic-Robin N, Dauplat MM, Mouret-Reynier MA, Van Praagh I, et al. Multicentric neoadjuvant pilot Phase II study of cetuximab combined with docetaxel in operable triple negative breast cancer. Int J Cancer 2016;138:2274–80. [DOI] [PubMed] [Google Scholar]
- 51.Finn RS, Press MF, Dering J, Arbushites M, Koehler M, Oliva C, et al. Estrogen receptor, progesterone receptor, human epidermal growth factor receptor 2 (HER2), and epidermal growth factor receptor expression and benefit from lapatinib in a randomized trial of paclitaxel with lapatinib or placebo as first-line treatment in HER2-negative or unknown metastatic breast cancer. J Clin Oncol 2009;27:3908–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Leo A, Gomez HL, Aziz Z, Zvirbule Z, Bines J, Arbushites MC, et al. Phase III, double-blind, randomized study comparing lapatinib plus paclitaxel with placebo plus paclitaxel as first-line treatment for metastatic breast cancer. J Clin Oncol 2008;26:5544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagaria TS, Shi C, Leduc C, Hoskin V, Sikdar S, Sangrar W, et al. Combined targeting of Raf and Mek synergistically inhibits tumorigenesis in triple negative breast cancer model systems. Oncotarget 2017;8:80804–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozma SC, Bogaard ME, Buser K, Saurer SM, Bos JL, Groner B, et al. The human c-Kirsten ras gene is activated by a novel mutation in codon 13 in the breast carcinoma cell line MDA-MB231. Nucleic Acids Res 1987;15:5963–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moasser MM, Basso A, Averbuch SD, Rosen N. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of HER2-overexpressing tumor cells. Cancer Res 2001;61:7184–8. [PubMed] [Google Scholar]
- 56.Brufsky A, Miles D, Zvirbule Z, Eniu A, Lopez-Miranda E, Seo J, et al. Abstract P5–21-01: Cobimetinib combined with paclitaxel as first-line treatment for patients with advanced triple-negative breast cancer (COLET study): Primary analysis of cohort I. Cancer Research 2018;78:P5–21-01. [Google Scholar]
- 57.Schmid P, Forster MD, Summers YJ, Good J, Sarker S-J, Lim L, et al. A study of vistusertib in combination with selumetinib in patients with advanced cancers: TORCMEK phase Ib results. Journal of Clinical Oncology 2017;35:2548. [Google Scholar]
- 58.Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med 2012;209:679–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carey JPW, Karakas C, Bui T, Chen X, Vijayaraghavan S, Zhao Y, et al. Synthetic Lethality of PARP Inhibitors in Combination with MYC Blockade Is Independent of BRCA Status in Triple-Negative Breast Cancer. Cancer Res 2018;78:742–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gravina GL, Festuccia C, Popov VM, Di Rocco A, Colapietro A, Sanita P, et al. c-Myc Sustains Transformed Phenotype and Promotes Radioresistance of Embryonal Rhabdomyosarcoma Cell Lines. Radiat Res 2016;185:411–22. [DOI] [PubMed] [Google Scholar]
- 61.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016;529:413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai L, Zhou B, Yang CY, Ji J, McEachern D, Przybranowski S, et al. Targeted Degradation of BET Proteins in Triple-Negative Breast Cancer. Cancer Res 2017;77:2476–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desrivieres S, Kunz C, Barash I, Vafaizadeh V, Borghouts C, Groner B. The biological functions of the versatile transcription factors STAT3 and STAT5 and new strategies for their targeted inhibition. J Mammary Gland Biol Neoplasia 2006;11:75–87. [DOI] [PubMed] [Google Scholar]
- 64.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest 2011;121:2723–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Muller U, et al. JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer. Cancer Cell 2012;22:796–811. [DOI] [PubMed] [Google Scholar]
- 66.Balko JM, Schwarz LJ, Luo N, Estrada MV, Giltnane JM, Davila-Gonzalez D, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 2016;8:334ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stover DG, Gil Del Alcazar CR, Brock J, Guo H, Overmoyer B, Balko J, et al. Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer 2018;4:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Locatelli MA, Aftimos P, Dees EC, LoRusso PM, Pegram MD, Awada A, et al. Phase I study of the gamma secretase inhibitor PF-03084014 in combination with docetaxel in patients with advanced triple-negative breast cancer. Oncotarget 2017;8:2320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu M, Peng Q, Jiang I, Carroll C, Han G, Rymer I, et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett 2013;328:261–70. [DOI] [PubMed] [Google Scholar]
- 70.Zhang CC, Yan Z, Zong Q, Fang DD, Painter C, Zhang Q, et al. Synergistic effect of the gamma-secretase inhibitor PF-03084014 and docetaxel in breast cancer models. Stem Cells Transl Med 2013;2:233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K, Zhang Q, Li D, Ching K, Zhang C, Zheng X, et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a gamma-secretase inhibitor. Clin Cancer Res 2015;21:1487–96. [DOI] [PubMed] [Google Scholar]
- 72.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 2011;17:1646–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 2012;486:395–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell 1999;4:511–8. [DOI] [PubMed] [Google Scholar]
- 75.Tutt A, Bertwistle D, Valentine J, Gabriel A, Swift S, Ross G, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 2001;20:4704–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 2000;275:23899–903. [DOI] [PubMed] [Google Scholar]
- 77.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917–21. [DOI] [PubMed] [Google Scholar]
- 78.Hartman AR, Kaldate RR, Sailer LM, Painter L, Grier CE, Endsley RR, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 2012;118:2787–95. [DOI] [PubMed] [Google Scholar]
- 79.Byrski T, Huzarski T, Dent R, Marczyk E, Jasiowka M, Gronwald J, et al. Pathologic complete response to neoadjuvant cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 2014;147:401–5. [DOI] [PubMed] [Google Scholar]
- 80.Hahnen E, Lederer B, Hauke J, Loibl S, Krober S, Schneeweiss A, et al. Germline Mutation Status, Pathological Complete Response, and Disease-Free Survival in Triple-Negative Breast Cancer: Secondary Analysis of the GeparSixto Randomized Clinical Trial. JAMA Oncol 2017;3:1378–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017 [DOI] [PubMed] [Google Scholar]
- 82.Litton J, Rugo H, Ettl J, Hurvitz S, Gonçalves A, Lee K-H, et al. Abstract GS6–07: EMBRACA: A phase 3 trial comparing talazoparib, an oral PARP inhibitor, to physician’s choice of therapy in patients with advanced breast cancer and a germline BRCA mutation. Cancer Research 2018;78:GS6–07-GS6-. [Google Scholar]
- 83.Wang Y, Krais JJ, Bernhardy AJ, Nicolas E, Cai KQ, Harrell MI, et al. RING domain-deficient BRCA1 promotes PARP inhibitor and platinum resistance. J Clin Invest 2016;126:3145–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Banda K, Swisher EM, Wu D, Pritchard CC, Gadi VK. Somatic Reversion of Germline BRCA2 Mutation Confers Resistance to Poly(ADP-ribose) Polymerase Inhibitor Therapy. JCO Precision Oncology 2018:1–6. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Bernhardy AJ, Cruz C, Krais JJ, Nacson J, Nicolas E, et al. The BRCA1-Delta11q Alternative Splice Isoform Bypasses Germline Mutations and Promotes Therapeutic Resistance to PARP Inhibition and Cisplatin. Cancer Res 2016;76:2778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pettitt SJ, Krastev DB, Brandsma I, Drean A, Song F, Aleksandrov R, et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat Commun 2018;9:1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ter Brugge P, Kristel P, van der Burg E, Boon U, de Maaker M, Lips E, et al. Mechanisms of Therapy Resistance in Patient-Derived Xenograft Models of BRCA1-Deficient Breast Cancer. J Natl Cancer Inst 2016;108. [DOI] [PubMed] [Google Scholar]
- 88.Stecklein SR, Kumaraswamy E, Behbod F, Wang W, Chaguturu V, Harlan-Williams LM, et al. BRCA1 and HSP90 cooperate in homologous and non-homologous DNA double-strand-break repair and G2/M checkpoint activation. Proc Natl Acad Sci U S A 2012;109:13650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu H, Di Antonio M, McKinney S, Mathew V, Ho B, O’Neil NJ, et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat Commun 2017;8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer 2004;4:814–9. [DOI] [PubMed] [Google Scholar]
- 91.Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 2003;95:1482–5. [DOI] [PubMed] [Google Scholar]
- 92.Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy With Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J Clin Oncol 2015;33:1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res 2016;22:3764–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Minckwitz GV, Timms K, Untch M, Elkin EP, Fasching PA, Schneeweiss A, et al. Prediction of pathological complete response (pCR) by Homologous Recombination Deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: Results from GeparSixto. Journal of Clinical Oncology 2015;33:1004-. [Google Scholar]
- 95.von Minckwitz G, Schneeweiss A, Loibl S, Salat C, Denkert C, Rezai M, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol 2014;15:747–56. [DOI] [PubMed] [Google Scholar]
- 96.Schrijver WA, Jiwa LS, van Diest PJ, Moelans CB. Promoter hypermethylation profiling of distant breast cancer metastases. Breast Cancer Res Treat 2015;151:41–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao EY, Shen Y, Pleasance E, Kasaian K, Leelakumari S, Jones M, et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res 2017;23:7521–30. [DOI] [PubMed] [Google Scholar]
- 99.Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP Inhibitor Upregulates PD-L1 Expression and Enhances Cancer-Associated Immunosuppression. Clin Cancer Res 2017;23:3711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nguyen M, Robillard L, Lin KK, Harding TC, Simmons AD. Abstract 1716: The PARP inhibitor rucaparib activates the STING pathway and enhances antitumor responses of immune checkpoint inhibitors in BRCA deficient syngeneic models. Cancer Research 2018;78:1716-. [Google Scholar]
- 101.Konstantinopoulos PA, Waggoner SE, Vidal GA, Mita MM, Fleming GF, Holloway RW, et al. TOPACIO/Keynote-162 (NCT02657889): A phase 1/2 study of niraparib + pembrolizumab in patients (pts) with advanced triple-negative breast cancer or recurrent ovarian cancer (ROC)—Results from ROC cohort. Journal of Clinical Oncology 2018;36:106-. [Google Scholar]
- 102.Roll JD, Rivenbark AG, Sandhu R, Parker JS, Jones WD, Carey LA, et al. Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol 2013;95:276–87. [DOI] [PubMed] [Google Scholar]
- 103.Kagara N, Huynh KT, Kuo C, Okano H, Sim MS, Elashoff D, et al. Epigenetic regulation of cancer stem cell genes in triple-negative breast cancer. Am J Pathol 2012;181:257–67. [DOI] [PubMed] [Google Scholar]
- 104.Karsli-Ceppioglu S, Dagdemir A, Judes G, Lebert A, Penault-Llorca F, Bignon YJ, et al. The Epigenetic Landscape of Promoter Genome-wide Analysis in Breast Cancer. Sci Rep 2017;7:6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Connolly RM, Li H, Jankowitz RC, Zhang Z, Rudek MA, Jeter SC, et al. Combination Epigenetic Therapy in Advanced Breast Cancer with 5-Azacitidine and Entinostat: A Phase II National Cancer Institute/Stand Up to Cancer Study. Clin Cancer Res 2017;23:2691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer 2012;12:465–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014;32:2959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010;28:105–13. [DOI] [PubMed] [Google Scholar]
- 109.Schmid P, Cruz C, Braiteh FS, Eder JP, Tolaney S, Kuter I, et al. Abstract 2986: Atezolizumab in metastatic TNBC (mTNBC): Long-term clinical outcomes and biomarker analyses. Cancer Research 2017;77:2986. [Google Scholar]
- 110.Dieci MV, Criscitiello C, Goubar A, Viale G, Conte P, Guarneri V, et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: a retrospective multicenter study. Ann Oncol 2014;25:611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miyashita M, Sasano H, Tamaki K, Hirakawa H, Takahashi Y, Nakagawa S, et al. Prognostic significance of tumor-infiltrating CD8+ and FOXP3+ lymphocytes in residual tumors and alterations in these parameters after neoadjuvant chemotherapy in triple-negative breast cancer: a retrospective multicenter study. Breast Cancer Res 2015;17:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol 2016;34:2460–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adams S, Schmid P, Rugo HS, Winer E, Loirat D, Cescon DW, et al. Phase 2 study of pembrolizumab (pembro) monotherapy for previously treated metastatic triple-negative breast cancer (mTNBC): KEYNOTE-086 cohort A. J Clin Oncol 2017;35:1088. [Google Scholar]
- 114.Adams S, Loi S, Toppmeyer D, Cescon D, De Laurentiis M, Nanda R, et al. Abstract PD6–10: KEYNOTE-086 cohort B: Pembrolizumab monotherapy for PD-L1–positive, previously untreated, metastatic triple-negative breast cancer (mTNBC). Cancer Research 2018;78:PD6–10. [Google Scholar]
- 115.Adams S, Diamond JR, Hamilton EP, Pohlmann PR, Tolaney SM, Molinero L, et al. Phase Ib trial of atezolizumab in combination with nab-paclitaxel in patients with metastatic triple-negative breast cancer (mTNBC). Journal of Clinical Oncology 2016;34:1009. [Google Scholar]
- 116.Tolaney S, Kalinsky K, Kaklamani V, Savulsky C, Olivo M, Aktan G, et al. Abstract PD6–13: Phase 1b/2 study to evaluate eribulin mesylate in combination with pembrolizumab in patients with metastatic triple-negative breast cancer. Cancer Research 2018;78:PD6–13. [Google Scholar]
- 117.Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, et al. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat 2018;167:671–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med 2018;379:2108–21. [DOI] [PubMed] [Google Scholar]
- 119.Emens L, Loi S, Rugo H, Schneeweiss A, Diéras V, Iwata H, et al. IMpassion130: Efficacy in immune biomarker subgroups from the global, randomized, double-blind, placebo-controlled, phase III study of atezolizumab + nab-paclitaxel in patients with treatment-naïve, locally advanced or metastatic triple-negative breast cancer. San Antonio Breast Cancer Symposium 2018 [Google Scholar]
- 120.Kwa MJ, Adams S. Checkpoint inhibitors in triple-negative breast cancer (TNBC): Where to go from here. Cancer 2018;124:2086–103. [DOI] [PubMed] [Google Scholar]
- 121.Barrett MT, Anderson KS, Lenkiewicz E, Andreozzi M, Cunliffe HE, Klassen CL, et al. Genomic amplification of 9p24.1 targeting JAK2, PD-L1, and PD-L2 is enriched in high-risk triple negative breast cancer. Oncotarget 2015;6:26483–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P, et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin Cancer Res 2016;22:1499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hellmann MD, Ciuleanu TE, Pluzanski A, Lee JS, Otterson GA, Audigier-Valette C, et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 2018;378:2093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saleh SMI, Bertos N, Gruosso T, Gigoux M, Souleimanova M, Zhao H, et al. Identification of Interacting Stromal Axes in Triple-Negative Breast Cancer. Cancer Res 2017;77:4673–83. [DOI] [PubMed] [Google Scholar]
- 125.Saleh S, Gruosso T, Gigoux M, Bertos N, Omeroglu A, Zuo D, et al. Abstract IA23: Deconvolution of the triple-negative breast cancer microenvironment. Cancer Research 2016;76:IA23–IA. [Google Scholar]
- 126.Harano K, Wang Y, Lim B, Seitz RS, Morris SW, Bailey DB, et al. Rates of immune cell infiltration in patients with triple-negative breast cancer by molecular subtype. PLoS One 2018;13:e0204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2018;379:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018;174:1373–87 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hoadley KA, Siegel MB, Kanchi KL, Miller CA, Ding L, Zhao W, et al. Tumor Evolution in Two Patients with Basal-like Breast Cancer: A Retrospective Genomics Study of Multiple Metastases. PLoS Med 2016;13:e1002174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Siegel MB, He X, Hoadley KA, Hoyle A, Pearce JB, Garrett AL, et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J Clin Invest 2018;128:1371–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 2017;32:169–84 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cejalvo JM, Martinez de Duenas E, Galvan P, Garcia-Recio S, Burgues Gasion O, Pare L, et al. Intrinsic Subtypes and Gene Expression Profiles in Primary and Metastatic Breast Cancer. Cancer Res 2017;77:2213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoefnagel LD, van de Vijver MJ, van Slooten HJ, Wesseling P, Wesseling J, Westenend PJ, et al. Receptor conversion in distant breast cancer metastases. Breast Cancer Res 2010;12:R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dieci MV, Barbieri E, Piacentini F, Ficarra G, Bettelli S, Dominici M, et al. Discordance in receptor status between primary and recurrent breast cancer has a prognostic impact: a single-institution analysis. Ann Oncol 2013;24:101–8. [DOI] [PubMed] [Google Scholar]
- 135.Chen T, Zhang N, Moran MS, Su P, Haffty BG, Yang Q. Borderline ER-Positive Primary Breast Cancer Gains No Significant Survival Benefit From Endocrine Therapy: A Systematic Review and Meta-Analysis. Clin Breast Cancer 2018;18:1–8. [DOI] [PubMed] [Google Scholar]
- 136.Iwamoto T, Booser D, Valero V, Murray JL, Koenig K, Esteva FJ, et al. Estrogen receptor (ER) mRNA and ER-related gene expression in breast cancers that are 1% to 10% ER-positive by immunohistochemistry. J Clin Oncol 2012;30:729–34. [DOI] [PubMed] [Google Scholar]
- 137.Symmans WF, Wei C, Gould R, Yu X, Zhang Y, Liu M, et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J Clin Oncol 2017;35:1049–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Masuda N, Lee SJ, Ohtani S, Im YH, Lee ES, Yokota I, et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N Engl J Med 2017;376:2147–59. [DOI] [PubMed] [Google Scholar]
- 139.Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 2014;6:514–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 2018;173:879–93 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gao R, Davis A, McDonald TO, Sei E, Shi X, Wang Y, et al. Punctuated copy number evolution and clonal stasis in triple-negative breast cancer. Nat Genet 2016;48:1119–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell 2016;164:293–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a Cancer Dependency Map. Cell 2017;170:564–76 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Witwicki RM, Ekram MB, Qiu X, Janiszewska M, Shu S, Kwon M, et al. TRPS1 Is a Lineage-Specific Transcriptional Dependency in Breast Cancer. Cell Rep 2018;25:1255–67 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stratikopoulos EE, Dendy M, Szabolcs M, Khaykin AJ, Lefebvre C, Zhou MM, et al. Kinase and BET Inhibitors Together Clamp Inhibition of PI3K Signaling and Overcome Resistance to Therapy. Cancer Cell 2015;27:837–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell 2015;163:174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016;167:260–74 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bardia A, Parton M, Kummel S, Estevez LG, Huang CS, Cortes J, et al. Paclitaxel With Inhibitor of Apoptosis Antagonist, LCL161, for Localized Triple-Negative Breast Cancer, Prospectively Stratified by Gene Signature in a Biomarker-Driven Neoadjuvant Trial. J Clin Oncol 2018:JCO2017748392. [DOI] [PubMed] [Google Scholar]
- 149.Hofmann D, Nitz U, Gluz O, Kates RE, Schinkoethe T, Staib P, et al. WSG ADAPT - adjuvant dynamic marker-adjusted personalized therapy trial optimizing risk assessment and therapy response prediction in early breast cancer: study protocol for a prospective, multi-center, controlled, non-blinded, randomized, investigator initiated phase II/III trial. Trials 2013;14:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]