Abstract
Anthrax is a lethal disease caused by the Gram-positive spore-producing bacterium Bacillus anthracis. We previously demonstrated that disruption of htrA gene, encoding the chaperone/protease HtrABA (High Temperature Requirement A of B. anthracis) results in significant virulence attenuation, despite unaffected ability of ΔhtrA strains (in which the htrA gene was deleted) to synthesize the key anthrax virulence factors: the exotoxins and capsule. B. anthracis ΔhtrA strains exhibited increased sensitivity to stress regimens as well as silencing of the secreted starvation-associated Neutral Protease A (NprA) and down-modulation of the bacterial S-layer. The virulence attenuation associated with disruption of the htrA gene was suggested to reflect the susceptibility of ΔhtrA mutated strains to stress insults encountered in the host indicating that HtrABA represents an important B. anthracis pathogenesis determinant. As all HtrA serine proteases, HtrABA exhibits a protease catalytic domain and a PDZ domain. In the present study we interrogated the relative impact of the proteolytic activity (mediated by the protease domain) and the PDZ domain (presumably necessary for the chaperone activity and/or interaction with substrates) on manifestation of phenotypic characteristics mediated by HtrABA. By inspecting the phenotype exhibited by ΔhtrA strains trans-complemented with either a wild-type, truncated (ΔPDZ), or non-proteolytic form (mutated in the catalytic serine residue) of HtrABA, as well as strains exhibiting modified chromosomal alleles, it is shown that (i) the proteolytic activity of HtrABA is essential for its N-terminal autolysis and subsequent release into the extracellular milieu, while the PDZ domain was dispensable for this process, (ii) the PDZ domain appeared to be dispensable for most of the functions related to stress resilience as well as involvement of HtrABA in assembly of the bacterial S-layer, (iii) conversely, the proteolytic activity but not the PDZ domain, appeared to be dispensable for the role of HtrABA in mediating up-regulation of the extracellular protease NprA under starvation stress, and finally (iv) in a murine model of anthrax, the HtrABA PDZ domain, was dispensable for manifestation of B. anthracis virulence. The unexpected dispensability of the PDZ domain may represent a unique characteristic of HtrABA amongst bacterial serine proteases of the HtrA family.
Keywords: Bacillus anthracis, HtrA protease, proteolytic domain, PDZ domain, auto-processing, virulence, stress sensitivity, anthrax
Introduction
Bacillus anthracis (B. anthracis), the etiological cause of the lethal anthrax disease is a spore-forming Gram-positive bacteria. In nature, B. anthracis exists as spores which exhibit notorious environmental resilience, and which are the infective form of the bacterium. The animals more frequently affected by the disease are wild or domesticated mammalian herbivores which contract the lethal spores while grazing. Upon infection of a host (cutaneous, gastro-intestinal or respiratory), the metabolically inert spores germinate into fast-dividing toxin-producing bacilli. Human cases of anthrax were frequent in the past due to contact with contaminated animal products or with carcasses of anthrax-succumbed animals. As of today, these cases are extremely rare in the Western world and the interest in the disease stems mainly from the potential intentional malicious use of B. anthracis spores as a bio-weapon (for reviews see Koehler, 2009; Chitlaru et al., 2011a).
The lethality of anthrax has been attributed to three main aspects of B. anthracis pathogenesis: the activity of the bacterial exotoxins, the anti-phagocytic role of its polyglutamate capsule and the remarkable proliferous nature of the bacteria in the host. This latter aspect of B. anthracis pathogenicity suggests that the pathogen excels in exploiting nutritional resources available in the host and is highly adapted to cope with stress constraints encountered in the course of infection. B. anthracis secretes two exotoxins, Lethal Toxin (LT) and Edema Toxin (ET) composed of binary combinations of the three proteins: Protective Antigen (PA), the non-harmful subunit of both toxins playing the essential role of binding to a receptor on the surface of host target cells and mediating the intracellular translocation of the lethal subunits of the toxin complex, Lethal Factor (LF), a zinc protease which together with PA forms the exotoxin LT (Klimpel et al., 1994) and Edema Factor (EF) an adenylate cyclase which together with PA constitutes the exotoxin ET, (Leppla, 1982; Lacy and Collier, 2002; Collier, 2009). The three components of the toxin, are encoded by genes located on pXO1, one of the two virulence plasmids naturally harbored by B. anthracis. A second well-established virulence factor is represented by a polyglutamate anti-phagocytic capsule synthesized by enzymes encoded by genes located on the second native plasmid pXO2. Anthrax is acknowledged as a toxinogenic disease, owing to the lethality of pure toxin preparations and pivotal role of the toxins in B. anthracis virulence, yet, during infection, B. anthracis secretes a large number of proteins, many of which bear biological functions indicative of a role in the onset and progression of the disease (Ariel et al., 2002, 2003; Chitlaru et al., 2006, 2007). As of today, a number of proteins, other than the classic toxins, have been suggested to play an essential role during B. anthracis infection, based on the attenuated virulence of null mutants entailing targeted disruption of specific genes (Chitlaru et al., 2011a; see Chitlaru et al., 2016 for a list and discussion of reported B. anthracis attenuating mutations).
For all organisms, quality control of protein synthesis is a vital activity. One central player in the context of protein quality control is represented by the HtrA (High Temperature Requirement A) family of serine proteases, which are structurally and functionally conserved across a wide range of evolutionary distinct phylogenetic classes both in prokaryots and eukaryots (reviewed by Clausen et al., 2011; Hansen and Hilgenfeld, 2013; Skórko-Glonek et al., 2013; Backert et al., 2018). HtrA proteins exhibit the dual biological activities of chaperones and proteases (Pallen and Wren, 1997; Spiess et al., 1999; Clausen et al., 2002). HtrA proteins exhibit a characteristic structure (see Figure 1A), composed of an N-terminal serine protease domain and at least one C-terminal PDZ domain that recognizes substrates and in some cases activates the protease function (Wilken et al., 2004; Kim and Kim, 2005). The proteolytic domain entails, among other, the conserved catalytic serine residue whose integrity is essential for proteolytic activity of all serine proteases of the HtrA family. Previous studies evidenced that often the bacterial HtrA characteristic N terminal protease domain (often referred to as a trypsin domain) and the C terminal PDZ domains distinctly impact the proteolytic and chaperone activities of the protein resulting in distinct effects on the phenotypic characteristics of the bacteria (see for example Bæk et al., 2011). While these domains are present in HtrA orthologs of all bacteria (schematically described in Figure 1A), the mutual interactions between the domains affecting their specific activities, in particular the impact of the PDZ domain on the proteolytic activity and substrate specificity, seem to differ among various HtrAs (Kim and Kim, 2005; Clausen et al., 2011; Hansen and Hilgenfeld, 2013; Skórko-Glonek et al., 2013). In addition to the trypsin and the PDZ domains, bacterial HtrAs may exhibit N-terminal canonical Sec export-signal peptides (Tsirigotaki et al., 2017), trans-membrane domains (which often enable the membrane localization of the protein) and self-processing domains (which were identified in a number of HtrA and promote self-removal of the N-terminal region of the protein, e.g., Albrecht et al., 2018). In Escherichia coli and B. subtilis, the HtrA family of proteases are important for the survival of the bacteria under different stress regimens (Pallen and Wren, 1997; Clausen et al., 2002; Antelmann et al., 2003). In addition, in Gram-positive bacteria, the HtrA chaperones/proteases are closely associated with the SecA membrane-translocation machinery suggesting that their targets are constituted by secreted proteins (Tjalsma et al., 2004). In some cases, HtrA was invoked as being directly involved in the proteolytic processing or secretion of specific virulence-associated proteins such as SpeB and hemolysin in Streptococcus pyogenes (Lyon and Caparon, 2004; Cole et al., 2007), Bordetella pertussis toxin S1 (Vitikainen et al., 2005) and possibly adhesin P1 of Streptococcus mutans (Crowley et al., 2008). In other Gram-negative pathogens, such as Helicobacter pylori, HtrA was shown to facilitate virulence manifestation by direct proteolysis targeting host proteins such as E-cadherin (Hoy et al., 2010; Tegtmeyer et al., 2016).
FIGURE 1.

(A) Schematic representation of bacterial serine proteases of the HtrA family. The highly conserved domains of the HtrA serine proteases are indicated. Sig (TM)-signal sequence and trans-membrane domain; Trypsin-the proteolytic domain. Bacterial HtrA proteases may exhibit one or two PDZ domains. The export signal peptide, trypsin and unique PDZ domain of HtrABA as well as the H153, D183, and S255 residues forming the catalytic triad of the proteolytic domain were unambiguously identified by alignment with amino-acid sequences of 30 different bacterial serine proteases of the HtrA family using the T-coffee multiple sequence alignment program (Notredame et al., 2000; http://tcoffee.crg.cat). Similar alignments was previously documented (for example, Pallen and Wren, 1997; Kim and Kim, 2005; Singh et al., 2011). Boxed amino-acid sequence alignment: the high sequence conservation around the catalytic H, D, and S residues (shown in red with yellow background) is exemplified for six different bacterial HtrA serine proteases (hydrophobic residues are shown in yellow; polar non-charged residues are shown in green; positively charged residues are shown in blue; negatively charged residues are shown in pink). The respective bacteria are indicated on the left of the alignment. (B) Schematic description of the three forms of HtrABA used in trans-complementation experiments for determining the role of the PDZ domain and the proteolytic activity of HtrABA: full-length (FL), truncated (ΔPDZ) or mutated (htrAS255A). The trans-complementation HtrABA forms were engineered to exhibit a C terminal stretch of 6 histidine residues (6 × His, brown box) enabling zinc-affinity purification and Western blot visualization using anti-His antibodies.
High through-put genomic/proteomic/serologic surveys of B. anthracis (reviewed in Chitlaru and Shafferman, 2009; Shafferman et al., 2010; Chitlaru et al., 2011a) showed that HtrA belongs to a class of exposed immunogenic putative-vaccine candidates. B. anthracis HtrA (HtrABA), which is located both in the membrane as well as in the secretome of the bacteria, is encoded by the unique monocistronic htrA gene (NCBI locus tag BA6330 in the B. anthracis Ames-ancestor reference strain). HtrABA was shown to be present in the circulation of infected animals and consequently could serve as a potential anthrax early disease-biomarker (Sela-Abramovich et al., 2009). Disruption of the htrA gene in toxinogenic B. anthracis Vollum (pXO1+; pXO2+) or Sterne (pXO1+; pXO2-) strains resulted in a dramatic attenuation in the guinea pig, murine and rabbit models of anthrax (Chitlaru et al., 2011b, 2016). Previous studies of these htrA-disrupted B. anthracis strains (to be referred from hereon as ΔhtrA, Chitlaru et al., 2010, 2011b, 2016, 2017) have demonstrated that while abrogation of HtrABA synthesis does not affect the in vitro growth of the bacteria in culture, it results in a complex phenotype exhibiting the following characteristics: (i) in vitro increased sensitivity to stress (heat, oxidative, salt, and ethanol), (ii) failure to up-regulate the NprA secreted protease under nutritional deprivation, (iii) inability to assemble the typical B. anthracis S-layer (in strains cured of the pXO1 and pXO2 native plasmids), (iv) propagation delay in a macrophage-infection assay (suggested to reflect the increased sensitivity to oxidative stress). The significant attenuation of virulence of B. anthracis ΔhtrA strains suggested that HtrABA is essential for manifestation of B. anthracis pathogenicity and accordingly, represents a novel B. anthracis virulence determinant. Consequently, htrA-gene disruption in the non-capsular Sterne-strain served for the development of an efficacious and safe next-generation live attenuated anthrax spore-vaccine (Chitlaru et al., 2016, 2017). The role of HtrABA in promoting tolerance to various stress stimuli, similar to that observed in a variety of Gram-positive and negative bacteria (Skórko-Glonek et al., 2013; Backert et al., 2018) may represent the basis for the dramatic virulence-attenuation of the htrA disrupted bacteria, reflecting the inability of the mutated cells to cope with stress conditions encountered in the host. Of note, resilience to oxidative stress in the course of infection is considered to represent an important feature of pathogenic bacteria in general, and B. anthracis in particular (Welkos et al., 2011).
In the current report we inspect the contribution of the protease and PDZ-domains of B. anthracis HtrA to its function (as evidenced by the phenotype associated with its disruption). Accordingly, muteins exhibiting either deletion of the PDZ domain or mutational abrogation of the proteolytic activity were expressed in the ΔhtrA (htrA-disrupted) strains enabling assessment of their ability to trans-complement their defective phenotype. Additionally, B. anthracis mutated strains exhibiting chromosomal allele modifications altering either the proteolytic domain or deletion of the PDZ domain were generated. The data suggest that the proteolytic activity of HtrA is essential for its self-processing and subsequent export of HtrABA in to the extracellular milieu. The PDZ domain appears to be dispensable for this autolytic process. Furthermore, the PDZ domain was dispensable for most of the inspected functions related to stress resilience as well as for the involvement of HtrABA in the assembly of the bacterial S-layer. On the other hand, PDZ was essential for the HtrABA role in mediating up-regulation of the extracellular protease NprA under starvation stress. In a murine model of anthrax, it is shown that the HtrABA PDZ domain, is not essential, while the proteolytic activity is necessary for manifestation of B. anthracis virulence.
Materials and Methods
Bacterial Strains, Media and Growth Conditions
Bacillus anthracis strains used in this study are listed in Table 1. E. coli strains, are detailed in Supplementary Table S1. B. anthracis were cultivated at 37°C, in FAG broth (3.3% tryptone, 2% yeast extract, 0.74% NaCl, 0.4% KH2PO4, 0.8% Na2HPO4, 2% glycerol, pH 8), in Brain-Heart Infusion (BHI, DIFCO/Becton Dickinson) or in NBY low-nutrient content medium [0.8% (w/vol) Nutrient broth (Difco), 0.3% Yeast extract (Difco) and 0.5% Glucose]. E. coli were cultivated at 37°C, in Luria-Bertani (LB, Difco). LB-agar was used as solid broth. E. coli strains were used for plasmid construction. Antibiotic concentrations used for the selection in LB agar/broth were: for E. coli strains, ampicillin (Amp, 100 μg ml-1); for B. anthracis strains, Chloramphenicol (Cm, 5 μg ml-1), kanamycin (10 μg ml-1), and erythromycin (5 μg ml-1).
Table 1.
Bacillus anthracis strains used in the present study.
| B. anthracis Strain | Nomenclature throughout the article | Significance; Reference | |
|---|---|---|---|
| (1) | Sterne | Sterne | Toxinogenic Non-capsular (pXO1+, pXO2-); (Turnbull, 1991) |
| (2) | Sterne ΔhtrA | SΔhtrA | Sterne strain with a deleted htrA gene; (Chitlaru et al., 2016) |
| (3) | ΔVollum | ΔV | Non-toxinogenic and non-capsular plasmid-cured (pXO1-, pXO2-) derived from the ATCC14578 Vollum virulent strain; (Chitlaru et al., 2010, 2011b) |
| (4) | ΔVollumΔhtrA | ΔVΔhtrA | ΔV strain with a deleted htrA gene; (Chitlaru et al., 2010, 2011b) |
| (5) | ΔVollumhtrADPDZ | ΔVhtrADPDZ | ΔV strain containing an htrA allele with a deleted PDZ domain |
| (6) | ΔVollumhtrAS255A | ΔVhtrAS255A | ΔV strain containing an htrA allele with a S255A point mutation |
| (7) | Sterne ΔhtrA/HtrA | SΔhtrA/HtrA | Sterne ΔhtrA trans-complemented with a full-length htrA gene |
| (8) | Sterne ΔhtrA/HtrAΔPDZ | SΔhtrA/HtrAΔPDZ | Sterne ΔhtrA trans-complemented with an htrA gene lacking the PDZ domain |
| (9) | Sterne ΔhtrA/HtrAS255A | SΔhtrA/HtrAS255A | Sterne ΔhtrA trans-complemented with an htrA S255A gene |
| (10) | ΔVollumΔhtrA/HtrA | ΔVΔhtrA/HtrA | ΔVΔhtrA trans-complemented with a full-length htrA gene |
| (11) | ΔVollumΔhtrA/HtrAΔPDZ | ΔVΔhtrA/HtrAΔPDZ | ΔVΔhtrA trans-complemented with an htrA gene lacking the PDZ domain |
| (12) | ΔVollumΔhtrA/HtrAS255A | ΔVΔhtrA/HtrAS255A | ΔVΔhtrA trans-complemented with an htrA S255A gene |
Bacillus anthracis Sterne (strain 1) is a non-capsular vaccine strain from the IIBR collection. Strains 5–12 were generated in the current study. Strains 7–12 are trans-complemented strains in the back-ground of Sterne ΔhtrA (strains 7–9) or ΔVollumΔhtrA (strains 10–12). See also Figure 1B for schematic description of the experimental approach using the strains listed in the table.
Plasmid and Strain Construction
Plasmids and oligonucleotide primers used in this study are summarized in Supplementary Table S1. The oligonucleotide primers were designed according to the genomic sequence of B. anthracis Sterne strain. Point mutations were introduced using QuikChange site-directed mutagenesis kit (Statagene/Agilent). Prior to transformation into B. anthracis all plasmids were propagated in the methylation deficient E. coli strain dam-dcm-. B. anthracis cells were electrotransformed as described (Cohen et al., 2000). To express HtrABA as C-terminal His tag protein under amy promoter, the htrA complete gene was cloned as a SnaBI/BamHI-digest of PCR product (BA531 and BA819C primers), replacing the pagA gene in the previously described vector pASC-α (Cohen et al., 2000). Similary, a 900 bp segment encoding the N terminal 300 amino acids of HtrABA was cloned into pASC-α (BA531 and BA820C primers) to generate the ΔPDZ version of HtrABA. To generate a catalytic mutant of HtrABA, the conserved Ser255 (on the basis of alignment with the amino-acid sequences of a variety of HtrA proteases) was mutated to an alanine residue. Oligonucleotide-directed mutagenesis was performed using primers BA823 and BA824C on plasmid pASC-α/HtrA, to generate pASC-α/HtrAS255A. The vectors used for disruption of htrA gene by allelic replacement were previously described (Chitlaru et al., 2017). pEGS-N’HtrA was used to disrupt the C’ terminus of htrA gene. The plasmid was constructed in four steps: (i) The vector pEGS-cya (Supplementary Table S1, Levy et al., 2012) was digested with SpeI and NotI; (ii) a SpeI –NotI restriction fragment of the htrA N’terminus segment (900 bp) derived by PCR using BA827 and BA828C was cloned into the vector from step (i); (iii) the vector generated in step (ii) was digested with AscI and SpeI; (iv) a AscI- SpeI restriction fragment of the htrA 5′UTR segment (900 bp) derived by PCR using BA829 and BA830C was cloned into the vector from step iii. pEGS-HtrAS255A was used to disrupt the conserved Ser255 of htrA gene. The plasmid was constructed in two steps: (i) The vector pEGS-cya was digested with AscI and NotI; (ii) a AscI –NotI restriction fragment of the htrAS255A derived by PCR using BA827 and BA831C and pASC-α/HtrAS255A as template was cloned into the vector from step (i).
Stress Sensitivity Agar Dilution Drop Assay
WT parental ΔV, ΔVΔhtrA and trans-complemented strains ΔVΔhtrA/HtrA, ΔVΔhtrA/HtrAΔPΔZ and ΔVΔhtrA/HtrAS255A (see Table 1 for nomenclature of strains used in this study and Supplementary Table S1 for the plasmids used to generate the trans-complemented strains), were grown in BHI to mid-log phase, brought to an OD of 1 OD unit and decimally diluted. All OD measurements were performed at a wavelength of 600 nm. Ten ml of each serial dilution was dropped on LB-agar plates containing various concentrations of freshly prepared H2O2 (final concentration 0.75 or 1.5 mM) or 3% NaCl. Plates were incubated at 37°C or at the indicated temperatures over-night.
DNA Preparation and Polymerase Chain Reaction (PCR)
Restriction enzymes (Fermentas) and T4 DNA ligase (Promega) were used as recommended by the supplier. Plasmid DNA were extracted from E. coli using Wizard Plus SV mini preps (Promega). PCR amplifications were performed using the MyTaq (Bioline) or Expand High Fidelity (Roche) systems. For fast colony screening, each colony was re-suspended in 25 μl PCR mix. PCR products were separated on 1% agarose gel using 1X TAE as running buffer and purified using QIAquick PCR purification kit (Qiagen). DNA sequences were determined with the ABI310 rhodamine termination reaction kit (ABI310 Genetic Analyzer, Applied Biosystems).
SDS-PAGE and Western Blot Analysis of B. anthracis Cultures
Bacterial pellets or bacterial secreted proteins (supernatants) collected from B. anthracis cultures [20 h post-inoculation with an over-night starter, at an initial optical density of 0.05 OD (optical density units), were analyzed by SDS-PAGE and Western blotting]. The final concentration of the culture was typically 10 OD (approx. 109CFU/ml) in Fag medium and 3 OD in NBY medium. Typically, the equivalent of 2 × 108 CFU (for analysis of the cell-associated fraction) or proteins secreted by 2 × 108 CFU (for analysis of the secreted material) was loaded per gel lane after 10 min boiling in SDS-load buffer. SDS-PAGE was carried out on 4–12% NuPage Bis-Tris gels (Invitrogen) using Precision Plus Molecular weight markers (Bio-Rad). Western blots were generated using the Nitrocellulose Western iBlot Gel transfer Semi-dry system (Invitrogen). The nitrocellulose membranes were blocked in LiCor blocking buffer for 1 h. at room temperature, and probed with primary antibody overnight at 4°C. The membranes were washed three times for 10 min in PBST (PBS containing 0.05% Tween), probed with secondary antibody for 1 h at room temperature and washed twice. The blots were scanned using the LiCor laser-based image detection method. The following antibodies were used in this study: mouse anti-HtrA (Chitlaru et al., 2011b), mouse anti-His (iii) rabbit anti-S layer (iv) mouse anti-NprA (v) IRDye®800CW conjugated goat anti-mouse (vi) IRDye®800CW conjugated goat anti-rabbit. Primary and secondary antibodies were used at 1:500 and 1:20,000 dilutions, respectively.
Expression in E. coli and IMAC (Immobilized Metal Affinity Chromatography) Purification of HtrA
To induce expression of HtrABA, an overnight culture in LB with 100 μg/ml ampicillin was diluted 1/100 in 20 ml fresh medium and grown to an optical density (600 nm) of 0.6; 1 mM IPTG (isopropyl-β-D-thiogalactopy-ranoside) was added and the culture continued for 4 h. Cells were harvested by centrifugation (10 min, 10,000 rpm, 4°C), washed once with PBS and pellet was frozen at -70°C until lysis. Cells were lysed on ice in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% tween20, pH 8.0) containing 1 mg/ml lysozyme, followed by sonication; lysates were cleared by centrifugation (30 min, 10,000g, 4°C). His-tagged HtrA proteins were purified by Ni-NTA magnetic agarose beads (Qiagene, 36111). Non-specifically bound proteins were removed with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 0.05% tween20, pH 8.0), and HtrA was eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.05% tween20, pH 8.0). Eluted proteins were dialyzed against PBS (Slide-A-lyzer 10K dialysis cassettes G2, #807729, Thermo Scientific).
Proteolytic Activity of HtrABA
Proteolytic activity of HtrABA, HtrABAΔPDZ or HtrAS255A was determined using casein as substrate as described (Bæk et al., 2011). In brief, reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 1 mg β-casein (Sigma C6905) and 0.4 μM HtrABA or HtrABAΔPDZ or HtrAS255A were incubated at 37°C for 18 h and terminated by the addition of 10 μl 4XSDS sample buffer and heating to 100°C for 5 min. samples were separated by SDS-PAGE and stained with GelCode blue stain reagent (Thermo scientific #24590).
Chaperone Activity of HtrA
Chaperone activity of HtrABA, HtrABAΔPDZ or HtrAS255A was performed under aggregation-prone conditions as described (Skórko-Glonek et al., 2003; Entzminger et al., 2012). In brief, egg-white lysozyme (Sigma L3790) was adjusted to 100 μM in denaturation buffer (HEPES-buffered saline, HBS [10 mM HEPES, 150 mM NaCl, pH 7.4] with 8 M urea and 50 mM DTT at pH 7.4) and allowed to equilibrate for 6 h. To monitor aggregation, unfolded and reduced lysozyme was rapidly diluted 100-fold into HBS alone or HBS containing HtrABA at concentration of 1 μM in 96-well plate. Protein aggregation was monitored by absorbance at 360 nm for up to 3 h at room temperature with a SpectroMax spectrophotometer.
Liquid Chromatography Mass Spectrometry (LC-MS) Mapping of HtrA
Liquid chromatography mass spectrometry of the heavy, light and truncated forms of HtrABA was performed at the De Botton Protein Profiling Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, Rehovot. In brief, gel bands containing the different forms of HtrABA were subjected to in-gel pepsin digestion. The resulting peptides were analyzed by nanoflow liquid chromatography coupled to high resolution, high mass accuracy mass spectrometry. The data was processed using Proteome Discoverer version 2.2.0.388, searched against the Uniprot Bacillus anthracis Ames ancestor reference strain protein database using both the SequestHT and Mascot search algorithms.
Infection of Experimental Animals
Outbred ICR mice (20–25 g, Harlan) were infected with vegetative cells of various B. anthracis Sterne-derived strains. All Sterne derived strains used in the virulence study, exhibited similar levels of LF, EF, and PA activity, determined by functional assays, as previously described (Israeli et al., 2016; Chitlaru et al., 2017). For infection, bacterial cultures were set in BHI medium by inoculation with an over-night starter at an initial density of 0.05 × OD units. Bacteria were grown for 3–4 h at 37°C, to mid-logarithmic phase, (approximately 2 × ODunits). Bacteria were inspected by microscope for similar chain length of vegetative cells, then centrifuged and re-suspended in PBS at the desired concentration (1 × OD unit = 108 CFU) such that mice were infected SC with 0.1 ml bacterial suspension and serial 10-fold dilutions. A total of 4 mice per dose per strain were used. The remaining bacterial dose suspensions were plated for total viable counts (CFU ml-1) to confirm the dose administered to the animals. For the virulence determination, the animals were observed daily for 21 days. The lethal dose required to kill 50% (LD50) of the animals was calculated by non-linear fit regression using the GraphPad Prism (version 5.0) statistical analysis software (San Diego, CA, United States). Animal experiments were approved by the IIBR committee for animal research. The IIBR animal-experiment protocol number was M-07-18. The experimental animals were handled according to the National Research Council 1996 Guide for the Care and Use of Laboratory Animals and regulations of the IIBR Animal Use Committee.
Results
General Design of the Study
To inspect the contribution of the protease and PDZ-domains of B. anthracis HtrA to its function, we generated trans-complementation vectors (Figure 1B) expressing three distinct recombinant forms of HtrABA, representing (i) the intact full-length of the protein (FL), (ii) a truncated form in which the PDZ domain is deleted (ΔPDZ), and (iii) an HtrABA form in which the catalytic serine necessary for proteolytic activity was point-mutated (S255A). The trans-complementation versions of HtrABA, tailored to express a C-terminal 6-his-tag were first expressed in E. coli, purified by immobilized metal affinity chromatography (IMAC) and used in assays which probed the proteolytic and chaperone activities, enabling confirmation that these activities are, as expected, differentially affected by the S255A mutation and by the PDZ deletion, respectively (Supplementary Figure S1).
The three forms of the proteins were then expressed in B. anthracis ΔhtrA, to interrogate the contribution of the protease and chaperone activities of HtrABA in determining the manifestation of major phenotypic features displayed by the ΔhtrA strains. In addition to this approach of trans-complementation of the ΔhtrA phenotype, an alternative, approach, consisting of direct modification of the chromosomal htrA allels, was carried out (Supplementary Figure S2). Accordingly, targeted gene-replacement modifications of the gene resulted in the generation of B. anthracis strains entailing total deletion of the htrA gene (ΔhtrA), partial deletion encompassing the PDZ domain (htrADPDZ) or point replacement mutation of the catalytic serine residue (htrAS255A). The various B. anthracis strains used in this study, exhibiting either allelic modified versions of the htrABA gene or expressing various trans-complementing forms of HtrABA in the background of the ΔhtrA strain are detailed in Table 1.
The Proteolytic Activity but Not the PDZ Domain Is Essential for Self-Processing and Consequent Secretion of HtrABA
Extrachromosomal expression of the full length (FL), ΔPDZ and S255A versions of HtrABA in B. anthracis ΔhtrA bacteria resulted in biosynthesis of the respective proteins which could be detected by Western-blot analysis in the cellular as well as secreted fractions, as depicted in Figure 2. Typically, the cellular fraction of B. anthracis WT cells contains 2 distinctly electrophoretic migrating HtrA bands corresponding to the FL and N-terminal processed protein (Figure 2), while the secreted fraction mainly contains the short processed form (Figure 2, see also Chitlaru et al., 2011b). The 2 forms will be referred hereon as H (heavy) and L (light). The detection of these two forms did not appear to reflect the forms of HtrA before and after the removal of the N-terminal signal peptide in the course of protein export (Tsirigotaki et al., 2017; Freudl, 2018), since the apparent electrophoretic migration-difference between the H and L forms implied the removal of an N-terminal fragment significantly longer than the predicted signal peptide (calculated to span the first 47 amino-acids of the protein, equivalent only to approximately 5 kDa) suggesting that the processing occurs at a location down-stream of the predicted signal-peptide cleavage site. Interestingly, the S255A mutated HtrABA migrated on SDS-gel as the H form (unprocessed) only. Furthermore, the mutated S255A version was much more abundant in the cellular fraction and almost undetected in its secreted form while the FL and ΔPDZ forms were abundantly present in the secretome (as well as in the cellular fraction). The fact that both anti-HtrA and anti-His antibodies generated similar Western-blot patterns (Figure 2) confirmed that the observed electrophoretic-migration differences stemmed from the processing of the N terminus (note that the His tag is located in the C terminus of the trans-complementation HtrA forms, see Figure 1B). These results establish that the proteolytic inactive HtrAS255A protein is retained in the bacteria and fails to be secreted probably due to its inability to process its N-terminus.
FIGURE 2.
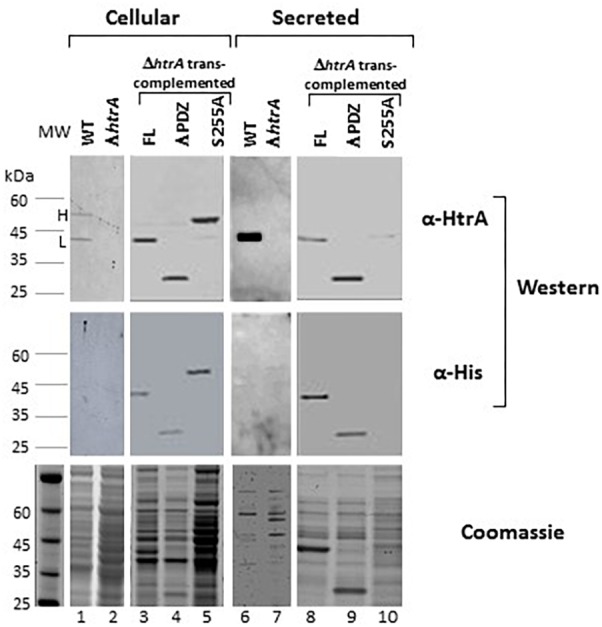
The S255A mutation abrogating the proteolytic activity prevents the N terminus auto-processing and secretion of HtrABA. Western-blot analysis of cellular (lanes 1–5) and secreted (lanes 6–10) proteins of the WT parental ΔVstrain (lanes 1 and 6), ΔVΔhtrA (lanes 2 and 7) and the trans-complemented strains ΔVΔhtrA/HtrA (lanes 3 and 8), ΔVΔhtrA/HtrAΔPDZ (lanes 4 and 9) and ΔVΔhtrA/HtrAS255A (lanes 5 and 10). Western blots were probed with anti-HtrA and anti-His antibodies, as indicated. The two forms of HtrABA are indicated as H (heavy, unprocessed) and light (auto-processed).
To further confirm that the auto-processing involves removal of the N-terminus and to map the site of the proteolysis, the electrophoretic distinct H and L forms of HtrABA, as well as the ΔPDZ version were inspected by liquid chromatography mass spectrometry (LC-MS) of in-gel pepsin digestion fragments (see section “Materials and Methods”). Digestion of the H and L forms established that peptides originating from the N-terminus of the protein could be detected only in the H form (representing the HtrAS255A mutant) while they were absent from the L form of HtrABA or the HtrABAΔPDZ version (Figure 3A). With the exception of the N-terminal region, digestion generated peptides covering most of the proteins. Comparison of the sequences of the N-terminal fragments detected in the H and L forms suggested that the cleavage of the N terminus occurred at position 103 of the protein (based on the detection of a pepsin fragment spanning amino acids 96–129 in the H form while the most N-terminal fragment detected in the L form spans amino acids 104–129). Cleavage at this site is in line with the difference (of about 10 kDa) observed between the electrophoretic migration of the two HtrABA forms (see detailed N-terminus sequence in Figure 3B). While we cannot accurately establish the exact site of the N-terminal self-proteolysis, it is conceivable that cleavage at this site results in deletion of a highly hydrophobic trans-membrane fragment whose removal is necessary for the release of the protein into the medium (squared fragment in Figure 3B). Of note, 3D structural modeling of HtrABA (using the PHYRE Protein Fold Recognition server, Kelley et al., 2015) established with high confidence that the first 100 amino acids of the protein constitute a highly exposed protruding domain (preceding the protease domain) which appears to be highly accessible to proteolysis without affecting the general architecture of the protein (Figure 3C). This observation supports the notion that the auto-processing occurs around position 100 of the protein.
FIGURE 3.

Liquid chromatography mass spectrometry (LC-MS) mapping of the heavy S255A (unprocessed), the light FL (auto-processed) and the truncated ΔPDZ forms of HtrABA. (A) The lines under the schematic linear depiction of HtrABA describe the position of the peptides detected by LC-MS in the respective forms of HtrABA collected from the SDS-gel as indicated by arrows. Positions of peptides obtained from digestion of the H form are indicated in blue while those from the L form or the ΔPDZ form are in red. (B) The amino-acid sequence of the first 120 residues of HtrABA. Brown residues are hydrophobic. The blue lines above the sequence indicate peptides identified by LC-MS in the heavy (H) form of HtrABAS255A. The red line above the sequence indicates the first peptide identified by LC-MS in the light (L) form of HtrABA. The highly hydrophobic trans-membrane domain is framed. The blue arrow indicates the predicted site of signal peptide cleavage. The red arrow indicates the predicted site of proteolytic auto-processing. (C) High confidence 3D structural modeling of HtrABA (created by using PHYRE Protein Fold Recognition modeling, Kelley et al., 2015) showing the clear spatial separation of the indicated protease and PDZ domains of the protein and that its first 100 amino acids constitute an exposed protruding domain (marked as N terminus), highly accessible to proteolysis without affecting the general architecture of the protein.
Taken together, these results provide support for the conclusion that (i) HtrABA is auto-processing its N-terminus as documented for a variety of bacterial HtrA paralogs (Skórko-Glonek et al., 1995, 2003; MohamedMohaideen et al., 2008; Hershey et al., 2016; Zhu et al., 2017; Albrecht et al., 2018, see section “Discussion”), (ii) that the proteolytic activity (abrogated by the S255A mutation) is essential for this process, (iii) that the N-terminus processing is necessary for efficient secretion of HtrABA, and (iv) finally and most intriguing, the PDZ domain is dispensable for the auto-proteolytic process.
The Proteolytic Activity but Not the PDZ Domain Is Essential for the Role of HtrABA in Growth of B. anthracis Under Temperature, Oxidative and Salt Stress
Involvement of HtrABA in the resilience of the bacteria to stress was demonstrated previously by determining the growth of ΔhtrA strains under various stress conditions and is considered to represent the main reason for the virulence attenuation associated with htrA gene disruption (Chitlaru et al., 2011b, 2016). To determine the relative contribution of the proteolytic or the PDZ domains to the role of HtrABA in the resilience of the bacteria to stress, ΔHtrA trans-complemented with the FL, ΔPDZ or S255A forms of HtrABA were subjected to a growth dilution drop-assay addressing sensitivity to high temperature, oxidative stress and salt, as depicted in Figure 4 (see also Supplementary Figure S3 for temperature sensitivity complementation using liquid cultures). The results establish that sensitivity to high temperature, hydroxide peroxide and salt exhibited by the htrA disrupted strain can be fully alleviated by trans-complementation with the FL or ΔPDZ forms but not with the S255A mutated form. The data support the conclusion that the proteolytic activity (abrogated by the S255A mutation) is essential, while the PDZ domain is dispensable for the role of HtrA in stress resilience.
FIGURE 4.
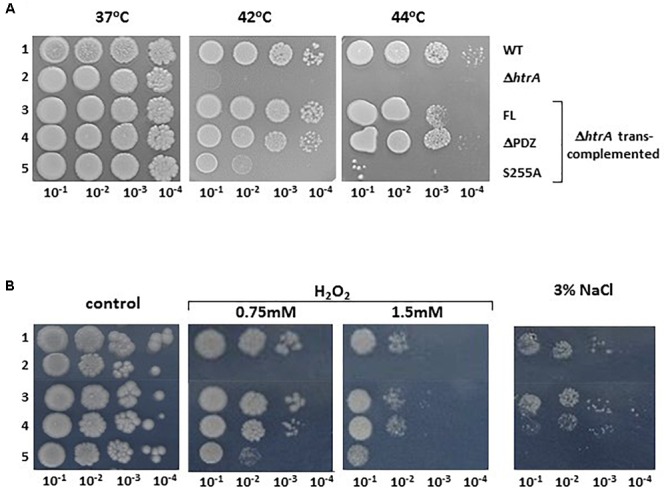
Dilution drop agar-assay of the WT parental ΔVstrain (1), ΔVΔhtrA (2), and the trans-complemented strains ΔVΔhtrA/HtrA (3), ΔVΔhtrA/HtrAΔPDZ (4) and ΔVΔhtrA/HtrAS255A (5) under various stress conditions. (A) Alleviation of the heat sensitivity phenotype of the ΔhtrA strain by trans-complementation with the FL and ΔPDZ forms of HtrABA. Plates were incubated at the indicated temperatures. (B) Alleviation of the H2O2 and NaCl sensitivity phenotype of the ΔhtrA strain by trans-complementation with the FL and ΔPDZ forms of HtrABA. Control LB-agar plates or LB plates containing the indicated concentrations of H2O2 or NaCl were incubated at 37°C.
The Proteolytic Activity but Not the PDZ Domain Is Essential for the Role of HtrABA in the Biosynthesis of B. anthracis S-Layer
We have documented in the past that in B. anthracis strains cured of the pXO1 and pXO2 native virulence-plasmids (such as the ΔVollum strain), disruption of the htrA gene results in the down-modulation of the B. anthracis S layer (Chitlaru et al., 2010, 2011b). This phenomenon, occurred post-transcriptionally and affected both the Sap and EA1 proteins which form the bacterial S-layer (Fouet, 2009) both in their cell-associated and secreted forms. Interestingly, this phenomenon was not observed in virulent B. anthracis strains (exhibiting the pXO1 virulence plasmid), suggesting that post-translational biosynthesis of the S-layer involves at least two alternative compensatory pathways, one governed by functions encoded in the pXO1 plasmid and another, HtrA-dependent mechanism which consists of chromosomally encoded functions (see Chitlaru et al., 2011b for an extensive discussion of this phenomenon). In the current study, we addressed the question whether the S-layer biosynthesis may be mediated by the mutated ΔPDZ or the S255A forms of HtrA. Accordingly, the level of S-layer proteins in the cell associated and secreted fractions of the wild-type parental (ΔVollum) as well as in the strains exhibiting chromosomally modified alleles ΔVΔhtrA, ΔVhtrADPDZ and ΔVhtrAS255A strains was evaluated by Western blot analysis using antibodies recognizing both the Sap and EA1 S-layer proteins. The data depicted in Figure 5A provide evidence that the ΔPDZ form of HtrA is sufficient to mediate the observed effect on S-layer modulation, similar to the effect observed in the parental strain, while the S255A non-proteolytic form of HtrA failed to assist the modulation of the S-layer bio-synthesis. This modulation of S-layer level by the FL or the ΔPDZ forms of HtrA (but not by the S255A mutant) observed in the chromosomally modified strains, was confirmed by the alternative trans-complementation approach of the ΔhtrA strain (Supplementary Figure S4). Based on the data obtained by both approaches it may be concluded that the proteolytic activity of HtrABA is essential for the S-layer biosynthesis modulation in the B. anthracis ΔVollum bacteria while the PDZ is dispensable for this role.
FIGURE 5.

Expression of S-layer proteins and of the NprA secreted protease by the B. anthracis parental WT ΔV, the ΔVΔhtrA and the ΔV strains exhibiting modified htrA alleles ΔVhtrADPDZ and ΔVhtrAS255A. (A) Western blot analysis of the secreted and cell associated protein fractions collected from BHI cultures of the strains using anti-S layer antibodies. (B) Western-blot analysis of the secreted protein fraction collected from low-nutrient NBY cultures of the strains using anti-NprA antibodies.
The PDZ Domain but Not the Proteolytic Activity Is Essential for the Role of HtrABA in Up-Regulation of the NprA Secreted Protease Under Low-Nutrient Conditions
In low-nutrient media, a prevalent part of the B. anthracis secretome is constituted by the secreted protease NprA, which is not expressed in high-nutrient culture and is down-regulated in CO2-enriched media (Chitlaru et al., 2006, 2010). The dramatic up-regulation of NprA has been suggested to reflect a response to the depletion of nutrients and consequent requirement for proteolytic activities enabling compensatory exploitation of proteins for survival. We have documented that ΔhtrA strains do not exhibit up-regulation of the NprA protease in low-nutrient media or following extended (>30 h) culture in rich media (Chitlaru et al., 2010, 2011b). This characteristic of the ΔhtrA strains was attributed to the role of HtrA in the ability of B. anthracis to cope with various stress regimens. The involvement of HtrA in the up-regulation of NprA involved induction of the nprA gene transcription, as opposed to the effect of HtrA on S-layer modulation, which occurs post-transcriptionally (see above). In the current study, we addressed the question whether the induction of NprA may be mediated by the mutated ΔPDZ or the S255A forms of HtrABA. Accordingly, the level of NprA in the low-nutrient cultures of the wild-type parental (ΔVollum), as well as the chromosomally modified ΔV ΔhtrA, ΔVhtrAΔPDZ and ΔVhtrAS255A strains was evaluated by Western blot analysis using anti NprA antibodies. The data depicted in Figure 5B provide evidence that the S255A non-proteolytic form of HtrA is sufficient to mediate the observed induction of NprA, while the bacteria expressing the ΔPDZ form of HtrABA did not exhibit induction of NprA. Thus, it may be concluded that the PDZ domain is essential for the induction of NprA in B. anthracis while the proteolytic activity of HtrA is dispensable for this role.
The Proteolytic Activity but Not the PDZ Domain Is Essential for the Role of HtrABA in Manifestation of B. anthracis Virulence
HtrA plays an essential role in B. anthracis virulence (Chitlaru et al., 2011b, 2016, 2017), as demonstrated by the observation that ΔhtrA strains are significantly less virulent than their isogenic parental strains. Disruption of the htrA gene results in attenuation of virulence unmatched by any mutation in B. anthracis, other than those affecting the bacterial toxins LT and ET (Chitlaru et al., 2016), in all rodent animal models of anthrax. This impact of htrA gene disruption on virulence, underlines the importance for anthrax pathogenesis of the ability of the bacteria to cope with the hostile environment, encountered in the infected host (see section “Introduction”). In the current study, by using an SC murine infection assay of B. anthracis virulence (Chand et al., 2009), we probed the importance of the protease and PDZ domains in enabling HtrABA to exert its role in B. anthracis virulence. While the model of choice for B. anthracis Sterne-strain infection is constituted by respiratory exposure of mice (or guinea pigs) to spores, sub-cutaneous administration of vegetative cells to mice provides a virulence model exhibiting an extended window of infection doses. Consequently, the ICR-outbred murine model of SC exposure was adequate for evaluation of the impact that various genetic manipulations and/or heterologous gene expression (such as trans-complemented strains) on the virulence of various strains (see for example Chitlaru et al., 2016, 2017). Thus, B. anthracis Sterne ΔhtrA cells trans-complemented with the FL, ΔPDZ or S255A forms of HtrABA were used side-by-side with the WT Sterne and the Sterne ΔhtrA strains for determination of their LD50 values in mice (Figure 6). All animals exposed to the Sterne strain succumbed 3–6 days post inoculation (LD50 = 10 CFU) while significant survival was demonstrated by the animals inoculated with the Sterne ΔhtrA strain (LD50 = 103 CFU), in line with previous experiments in the murine system (Chitlaru et al., 2016). As expected, trans-complementation with the FL form of HtrA fully restored the virulence of the bacteria, as attested by an LD50 value of 10 CFU, undistinguishable of that exhibited by the WT parental Sterne strain. Interestingly, the same full complementation effect was mediated by expression of the ΔPDZ form of HtrA, but not by expression of the S255A form (Figure 6). Thus, it may be concluded that the proteolytic activity of HtrA is essential for B. anthracis virulence while the PDZ is dispensable for its role.
FIGURE 6.

Virulence of B. anthracis Sterne, SΔhtrA, and the trans-complemented strains SΔhtrA/HtrA, SΔhtrA/HtrAΔPDZ, and SΔhtrA/HtrAS255A, in a murine model of virulence assessment. (A) Survival table of mice infected by sub-cutaneous administration of the various strains. Each experimental group included 4 mice. The resulting Lethal Dose 50% (LD50) and the 95% confidence intervals were calculated by global non-linear fit regression using the GraphPad Prism software. The statistical significance of the calculated LD50 values was determined by the extra-sum-of-squares F comparison test using the GraphPad Prism software; ∗p < 0.05. (B) Kaplan–Meier survival chart of selected experimental groups, as indicated in the boxed legend. Note that 104 bacteria of the SΔhtrA or SΔhtrA/HtrAS255A strains were administrated to mice, underlying the virulence attenuation of these strains; only 102 SΔhtrA/HtrAΔPDZ bacteria were administered underlying the high virulence exhibited by the SΔhtrA strain upon trans-complementation with the ΔPDZ form of HtrABA.
Discussion
HtrABA is an important virulence determinant of B. anthracis (Chitlaru et al., 2011b). Owing to the very high virulence attenuation associated with its disruption (6 orders of magnitude increase in the LD50 value compared to the parental strain in the guinea pig model, Chitlaru et al., 2016), which does not affect the synthesis and secretion of the B. anthracis Protective Antigen (PA, necessary for the elicitation of protective immunity), B. anthracis ΔhtrA strains were recently shown to represent efficacious next generation live attenuated anthrax vaccine (Chitlaru et al., 2016, 2017). It is interesting to note that HtrABA is a unique mono-cistronic gene, unlike the situation in B. subtilis which possess 2 highly homologous genes, HtrA (the product of the ykdA gene) and HtrB (the product yvtA gene) which may mutually compensate when the function of one of them is abrogated (Hyyrylainen et al., 2001; Noone et al., 2001; Darmon et al., 2002). Such a compensatory mechanism is not active in B. anthracis, therefore disruption of HtrABA is associated with a well-defined phenotype. Yet, the B. anthracis ΔhtrA strains are not affected in their in vitro growth and appear to manifest their phenotype in conditions which are reminiscent of the in vivo infection (Chitlaru et al., 2011b). As many other bacterial HtrA paralogs, HtrABA is composed of a signal peptide, trans-membrane domain, a trypsin-like protease catalytic domain, and a C-terminal PDZ domain. In the study documented in the current report we interrogated the relative impact that the proteolytic activity (mediated by the catalytic trypsin-like protease domain) and the PDZ domain (necessary for the chaperone activity) have in manifestation of the various functions mediated by HtrABA. Based on trans-complementation of ΔhtrA strains with either a wild-type, truncated (ΔPDZ), or non-proteolytic form (S255A) of HtrABA, or by inspection of strains exhibiting modified chromosomal alleles of HtrABA, it is shown that the proteolytic activity is necessary for the N-terminus processing and subsequent secretion of HtrABA, for the role of HtrABA in resilience of the bacteria to heat, oxidative and salt stress, for the role in the biosynthesis of the S-layer and is essential for manifestation of the role of HtrABA in virulence, while the PDZ domain is dispensable for all these roles. On the other hand, the proteolytic activity appears to be dispensable for the HtrA-mediated induction of the NprA protease under limited nutrient conditions.
Self-Processing and Secretion of HtrABA
One of the major observations of this study is that proteolytic active forms of HtrABA (either the full-length or the ΔPDZ versions) undergo N-terminal proteolytic self-processing which results in the removal of a highly hydrophobic putative trans-membrane domain and consequently in the release of HtrABA as a secreted protein (Figure 2). By LC-MS peptide mapping of the autolytic processed and un-processed forms of HtrABA, we were able to map the putative N-terminal cleavage site to the methionine residue at position 103. Removal of the N-terminal domain of HtrABA by self-processing in the course of its secretion is suggested also by the recent observation of Pomerantsev et al. (2017) who documented that a shorter than expected form of over-expressed HtrA was released from an engineered B. anthracis protease-free strain (exhibiting deletion of 10 various proteases). The boundary of the removed N-terminal segment was mapped at position 76 of the protein, about 23 amino acids up-stream of the cleavage site implied by our data. This inconsistency may stem from the differences in HtrA expression level of the systems used in the two studies or (alternatively but not mutually exclusive) from the fact that different B. anthracis strains were used in the two studies; interestingly, one simple interpretation which may reconcile the discrepancy is that an additional proteolytic activity absent from the Ames BH500 non-proteolytic engineered strain (Pomerantsev et al., 2011, 2017) but present in the ΔVollum strain employed in the current study, may be involved in the processing of the N terminus of HtrA, in conjunction with the autolytic activity. In any case, while this issue awaits to be resolved, both studies bring evidence that the removal of a highly hydrophobic N-terminal region, downstream of the canonical export signal sequence, is an essential step for the secretion of HtrA.
The mechanism by which HtrABA undergoes both removal of the N-terminal canonical export-signal peptide and autolytic processing of the N-terminus represents a matter of further study. Interestingly, the autolytic unprocessed HtrABa (which prevails in the case of the proteolytically deficient S255A mutant and consequently fails to be secreted, Figure 2) harbors an intact N-terminus, indicating that its signal peptide was not removed (see LC-MS mapping peptide coverage of HtrABA in Figure 3A,B). The fact that the S255A mutation prevents not only the N terminus autolytic processing but also the removal of the HtrABA canonical Sec-pathway signal peptide by a putative peptidase (Auclair et al., 2012; Tsirigotaki et al., 2017) may indicate that the two processes occur in concert or that they are mechanistically connected. Theoretically, the two processes are redundant and independent, and removal of the signal peptide by a putative peptidase may have been sufficient to disrupt the hydrophobic domain and consequently to release the protein from its membrane form. Therefore it is possible that N-terminal self-processing may be important for other aspects of HtrA function, in addition to its secretion.
N-terminal autolytic cleavage of HtrA has been observed in several other serine proteases of the HtrA family. The E. coli DegP serine protease undergoes N-terminal autolytic degradation (Skórko-Glonek et al., 1995) provided that its proteolytic catalytic triad is intact. In the case of DegP, the autolytic process appeared to be mainly necessary for the control and modulation of the quaternary/oligomeric structure of the protein, rather than for its secretion: N terminus self-proteolysis of DegP results in the removal of two cysteine residues which are involved in stabilization and multimerization of the molecules via disulfide bridges (Skórko-Glonek et al., 2003). Of note, B. anthracis HtrA N-terminus does not contain any cysteine residue and furthermore, it does not appear to be involved in determining the general architecture of the monomeric molecule (see Figure 3C). The Mycobacterium tuberculosis HtrA2 protein (Mtb HtrA2), one of the three HtrA-like serine proteases harbored by this bacterium, was also suggested to be released into the medium via an autolytic process, yet in this case, the protein does not exhibit a recognizable secretion peptide sequence (MohamedMohaideen et al., 2008). Furthermore, the autolytic process of Mtb HtrA2 targets additional sites resulting in the generation of short regulatory peptides which are incorporated in the binding cavities of the proteolytic and PDZ domains of the molecule. A similar situation does not seem to occur in HtrABA, based on the observation that no additional proteolytic products of the protein were detected by Western blot analysis. An additional case of amino-terminal self-processing of HtrA was recently reported in H. pylori (Albrecht et al., 2018). H. pylori HtrA (HbP HtrA) is considered to play an important role in virulence, attributed to its proteolytic activity, specifically targeting host E-cadherin (Hoy et al., 2010). Subsequently, this HtrA-mediated mechanism of virulence was demonstrated in additional Gram-negative gastrointestinal pathogens (Hoy et al., 2012; Abfalter et al., 2016). Autolysis of HbP HtrA appears to have a dual role: on the one hand it is necessary for secretion of the protein. On the other hand, N-terminus autolytic processing affects cleavage of the E-cadherin substrate by modulating the oligomerization state of membrane-bound HbP HtrA which is essential for its proteolytic activity. Recently, the HtrA-like protease from the thermophilic Brevibacillus sp. WF146 strain was shown to be released from the cell surface via autoprocessing of its N-terminal membrane anchor (Zhu et al., 2017), yet interestingly, in this case the autoprocessing required the association of the PDZ domain of the protein with a denatured proteolysis substrate. A similar dependence of the HtrA autolytic process to association of the PDZ domain with substrates was demonstrated in Magnetospirillum magneticum HtrA-like protein MamE (Hershey et al., 2016), yet in this case the issue of secretion was not addressed. A similar mechanism linking substrate recognition to auto-proteolysis does not appear to occur in B. anthracis in which the N-terminal processing occurs efficiently with truncated ΔPDZ forms of HtrABA (Figure 2 and see below). Taken together, these studies indicate that N-terminal self-processing is a common characteristic of HtrA serine proteases yet the role of self-processing may differ from case to case. We propose that in the case of HtrABA, the major role of N-terminal autolysis is secretion of the protein. In line with this concept, we have documented in the past that HtrABA is abundantly secreted in vivo (coinciding with expression of the anthrax toxins), can be detected in the circulation of infected animals and consequently can serve as an early biomarker for B. anthracis infection (Chitlaru et al., 2006; Sela-Abramovich et al., 2009).
The Proteolytic Activity of HtrABA Is Essential While Its PDZ Domain Appears to Be Dispensable for Most of Its Functions
The proteolytic activity of HtrABA, abrogated by the S255A mutation, is demonstrated to be essential not only for autolytic processing, as explained above, but also for the role of HtrABA in heat, oxidative, and salt resilience (Figure 3 and Supplementary Figure S3), for its role in the biosynthesis of S-layer (Figure 5A and Supplementary Figure S4) and finally for its role in manifestation of B. anthracis virulence (Figure 6). These conclusions are based on the failure of the HtrA S255A form to trans-complement the phenotype exhibited by the ΔhtrA strain. Actually, the only iteration in which the role of HtrA was not affected by the S255A mutation (but rather by the PDZ domain deletion) was induction of the NprA extracellular protease (Figure 5B). This former aspect of HtrA function involves the (direct or indirect) role of HtrA as a stress-related regulatory factor (Chitlaru et al., 2011b), which is not related to its proteolytic ability but rather to a more pleiotropic role of HtrABA in modulating expression of genes involved in stress response. Accordingly an extensive transcriptomic analysis of HtrABA mutated strains evidenced HtrA-dependent expression-regulation of a large number of genes under various stress regimens (manuscript in preparation).
The essentiality of the proteolytic activity is a hallmark of all serine proteases of the HtrA family (extensively reviewed by Pallen and Wren, 1997; Kim and Kim, 2005; Clausen et al., 2011; Singh et al., 2011; Hansen and Hilgenfeld, 2013; Skórko-Glonek et al., 2013; Chang, 2016; Backert et al., 2018). In this regard, HtrABA is not an exception. Yet the dependence on HtrA activity on the integrity of the proteolytic domain is not always as strict as evidenced in this study. For example, in Campylobacter jejuni (Bæk et al., 2011), HtrA chaperone activity is sufficient for growth under mild stress conditions. Furthermore, proteolytic inactive forms of E. coli DegP (exhibiting an S210A mutation, analogous to the S255A of HtrABA employed in the current study) were efficient in trans-complementing the temperature sensitivity of an htrA mutant strain (Skórko-Glonek et al., 2007). These studies suggested that sometimes association of HtrA to misfolded proteins without their degradation may be sufficient for maintenance of stress resilience. This appears not to be the case with HtrABA in which stress sensitivity of the ΔhtrA strain could not be alleviated by expression of the S255A variant of HtrABA. As mentioned above, the proteolytic-inactive S255A form of HtrABA fails to be secreted, therefore additional studies will be required to determine whether the failure of this form to trans-complement the ΔhtrA stress-sensitivity phenotype is due to the loss of proteolytic activity per se or as a result of its intracellular retention. Yet, it is conceivable that the role of HtrABA in stress resilience involves catalytic activities performed intracellularly, such as degradation of mal-folded proteins accumulating under stress, therefore it is not likely that this trans-complementation failure can be attributed to its intracellular retention. Furthermore, we note that, in preliminary experiments, the virulence attenuation of the ΔhtrA strain could not be alleviated in trans by co-infection with an HtrA overproducing strain (data not shown) suggesting that secretion of HtrA is not essential for manifestation of its role in virulence.
While the importance of the HtrABA proteolytic domain, was expected considering the fact that the major role of HtrA is related to the quality control of proteins and degradation of mal-folded proteins which may accumulate under stress conditions, our data show, that the PDZ domain of HtrABA is dispensable for many of its functions (see below) and most importantly for manifestation of the B. anthracis virulence. This observation is presumably, the most unexpected out-come of this study and is most-intriguing in light of the extensive studies carried out with similar proteins which demonstrated the pivotal role of the PDZ domain for their function. For example, in contrast to our observations with HtrABA, studies of various mutated variants of Salmonella enterica (S. Typhimurium) HtrA established that absence of either one of the 2 PDZ domains present in this protein completely abolished the ability of HtrA to complement the growth defects of an htrA mutant (Lewis et al., 2009). A similar situation was reported in Legionella pneumophila HtrA, although in this case, trans-complementation of the intracellular replication defect of an HtrA mutant, could be partially promoted by a truncated variant containing only the first PDZ domain and completely abolished only by deletion of both PDZ domains (Pedersen et al., 2001). Analogous observations supporting the essentiality of the PDZ domain were documented in E. coli DegP in which deletion of its both PDZ domains completely abolished the protease activity of the protein and expression of such a truncated form in an htrA mutant could not complement its temperature sensitive phenotype (Spiess et al., 1999). Similarly to Legionella HtrA, only the first PDZ domain of DegP emerged as absolutely essential for the activity of HtrA (Iwanczyk et al., 2007).
It is possible that the dispensability of the PDZ domain of HtrABA reflects an inhibitory role of the PDZ domain on the proteolytic activity, such as suggested for DegS, the E. coli stress response protease and an archetype of bacterial serine proteases of the HtrA family. DegS was shown to exist in a proteolytic-inactive form in which the PDZ domain allosterically prevents the activity of the protease domain by restricting the access to the protease active site (Wilken et al., 2004). The PDZ domain was demonstrated to bind peptides derived from mal-folded proteins accumulating during hostile environmental conditions, and consequently to relieve the allosteric inhibition resulting in activation of the proteolytic activity (Walsh et al., 2003; Wilken et al., 2004). If that was the case also in HtrABA, the deletion of PDZ domain would cause perpetuation of the proteolytic active conformation resulting in the ability of the ΔPDZ truncated form to trans-complement the ΔhtrA phenotype, as indeed recorded in our study. Yet, the analogy between DegS and HtrABA is only limited, since, unlike the situation in B. anthracis, a truncated form of DegS lacking the PDZ domain exhibited only marginal levels of proteolytic activity (Walsh et al., 2003) suggesting that the DegS PDZ domain may have additional positive roles on the overall activity of DegS. It is interesting to note that a similar dispensability of the PDZ domain has been reported only for the human proteases HtrA1 and HtrA3, representing eukaryotic paralogs of bacterial quality-control serine proteases of the HtrA family, in which it appears that activity is not affected by deletion of their respective PDZ domains (Truebestein et al., 2011; Glaza et al., 2015).
Conclusion
Here we presented for the first time a study addressing the importance of the proteolytic and PDZ domains of B. anthracis HtrA for its function and most importantly for manifestation of B. anthracis virulence. These data strongly suggest that mutational abrogation of the proteolytic activity of HtrA and truncation of its PDZ domain, have different consequences on the activity of the protein. The major conclusions of the study are (i) the proteolytic activity of HtrABA is essential for its autolysis and secretion, for the role of HtrA in stress resistance and for manifestation of B. anthracis virulence, and (ii) the unique PDZ domain is dispensable for all these processes. Accordingly, HtrABA exhibits significant differences compared to other bacterial serine proteases of the HtrA family. The observations documented in this report are relevant to the design of novel therapeutic strategies targeting B. anthracis HtrA.
Author Contributions
TC conceptualized the study. MI, SR, UE, HC, TC, AB-K, and AT performed experiments. SR performed statistical analysis of data. TC, MI, and OC conceived, coordinated, and supervised the study. TC, AB-K, and OC analyzed and interpreted the data. TC wrote the manuscript. All authors revised and agreed on the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Yoseph Shlomovitch is thanked for his expert technical assistance in the animal experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00255/full#supplementary-material
References
- Abfalter C. M., Schubert M., Götz C., Schmidt T. P., Posselt G., Wessler S. (2016). HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal. 14:30. 10.1186/s12964-016-0153-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht N., Tegtmeyer N., Sticht H., Skorko-Glonec J., Backert S. (2018). Amino-terminal processing of Helicobacter pylori serine protease HtrA: role in oligomerization and activity regulation. Front. Microbiol. 9:642. 10.3389/fmicb.2018.00642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antelmann H., Darmon E., Noone D., Veening J., Westers H., Bron S., et al. (2003). The extracellular proteome of B. Subtilis under secretion stress conditions. Mol. Microbiol. 49 143–156. 10.1046/j.1365-2958.2003.03565.x [DOI] [PubMed] [Google Scholar]
- Ariel N., Zvi A., Grosfeld H., Gat O., Inbar Y., Velan B., et al. (2002). Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1: in silico and in vitro screening. Infect. Immun. 70 6817–6827. 10.1128/IAI.70.12.6817-6827.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel N., Zvi A., Makarova K. S., Chitlaru T., Elhanany E., Velan B., et al. (2003). Genome-based bioinformatic selection of chromosomal Bacillus anthracis putative vaccine candidates coupled with proteomic identification of surface-associated antigens. Infect. Immun. 71 4563–4579. 10.1128/IAI.71.8.4563-4579.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair S. M., Bhanu M. K., Kendal D. A. (2012). Signal peptidase I: cleaving the way to mature proteins. Protein Sci. 21 13–25. 10.1002/pro.757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S., Bernegger S., Skórko-Glonek J., Wessler S. (2018). Extracellular HtrA serine proteases: an emerging new strategy in bacterial pathogenesis. Cell. Microbiol. 20:e12845. 10.1111/cmi.12845 [DOI] [PubMed] [Google Scholar]
- Bæk K. T., Vegge C. S., Skórko-Glonek J., Brøndsted L. (2011). Different contributions of HtrA protease and chaperone activities to Campylobacter jejuni stress tolerance and physiology. Appl. Environ. Microbiol. 77 57–66. 10.1128/AEM.01603-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chand S. C., Drysdale M., Lovchhik J., Koehler T. M., Lipscomb M. F., Rick Lyons C. (2009). Discriminating virulence mechanisms among Bacillus anthracis strains by using a murine subcutaneous infection model. Infect. Immun. 77 429–435. 10.1128/IAI.00647-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z. (2016). The function of the DegP (HtrA) protein: protease versus chaperone. IUBMB Life 11 904–907. 10.1002/iub.1561 [DOI] [PubMed] [Google Scholar]
- Chitlaru T., Altboum Z., Reuveny S., Shafferman A. (2011a). Progress and novel strategies in vaccine development and treatment of anthrax. Immunol. Rev. 239 221–236. 10.1111/j.1600-065X.2010.00969.x [DOI] [PubMed] [Google Scholar]
- Chitlaru T., Zaide G., Ehrlich S., Inbar I., Cohen O., Shafferman A. (2011b). HtrA is a major virulence determinant of Bacillus anthracis. Mol. Microbiol. 81 1542–1559. 10.1111/j.1365-2958.2011.07790.x [DOI] [PubMed] [Google Scholar]
- Chitlaru T., Gat O., Gozlan Y., Ariel N., Shafferman A. (2006). Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 188 3551–3571. 10.1128/JB.188.10.3551-3571.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitlaru T., Gat O., Gozlan Y., Grosfeld H., Inbar I., Shafferman A. (2007). Identification of in vivo expressed immunogenic proteins by serological proteome analysis of Bacillus anthracis secretome. Infect. Immun. 75 2841–2852. 10.1128/IAI.02029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitlaru T., Gat O., Zaide G., Grosfeld H., Inbar I., Ehrlich S., et al. (2010). “Proteomic studies of B. anthracis reveal in-vitro CO2-Moldulation and expression during infection of extracellular proteases,” in The Challenge of Highly Pathogenic Microorganisms, Mechanisms of Virulence and Novel Medical Countermeasures eds Shafferman A., Velan B., Ordentlich A. (Netherland: Springer Press; ) 11–22. [Google Scholar]
- Chitlaru T., Israeli M., Bar-Haim E., Elia U., Rotem S., Ehrlich S., et al. (2016). Next-generation Bacillus anthracis live attenuated spore vaccine based on the htrA(-) (High Temperature Requirement A) Sterne Strain. Sci. Rep. 6:18908. 10.1038/srep18908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitlaru T., Israeli M., Rotem S., Elia U., Bar-Haim E., Ehrlich S., et al. (2017). A novel live attenuated anthrax spore vaccine based on an acapsular Bacillus anthracis Sterne strain with mutations in the htrA, lef and cya genes. Vaccine 35 6030–6040. 10.1016/j.vaccine.2017.03.033 [DOI] [PubMed] [Google Scholar]
- Chitlaru T., Shafferman A. (2009). Proteomic studies of Bacillus anthracis. Fut. Microbiol. 4 983–998. 10.2217/fmb.09.73 [DOI] [PubMed] [Google Scholar]
- Clausen T., Kaiser M., Huber R., Ehrmann M. (2011). HTRA proteases: regulated proteolysis in protein quality control. Nat. Rev. 12 152–162. 10.1038/nrm3065 [DOI] [PubMed] [Google Scholar]
- Clausen T., Southan C., Ehrmann M. (2002). The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10 443–455. 10.1016/S1097-2765(02)00658-5 [DOI] [PubMed] [Google Scholar]
- Cohen S., Mendelson I., Altboum Z., Kobiler D., Elhanany E., Bino T., et al. (2000). Attenuated nontoxinogenic and nonencapsulated recombinant Bacillus anthracis spore vaccines protect against anthrax. Infect. Immun. 68 4549–4558. 10.1128/IAI.68.8.4549-4558.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N. C., Aquilina J. A., Hains P. G., Henningham A., Sriprakash K. S., Caparon M. G., et al. (2007). Role of group A Streptococcus HtrA in the maturation of SpeB protease. Proteomics 7 4488–4498. 10.1002/pmic.200700626 [DOI] [PubMed] [Google Scholar]
- Collier R. J. (2009). Membrane translocation by anthrax toxin. Mol. Aspects. Med. 30 413–422. 10.1016/j.mam.2009.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley P. J., Seifert T. B., Isoda R., van Tilburg M., Oli M. W., Robinette R. A., et al. (2008). Requirements for surface expression and function of Adhesin P1 from Streptococcus mutans. Infect. Immun. 76 2456–2468. 10.1128/IAI.01315-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmon E., Noone D., Masson A., Bron S., Kuipers O. P., Devine K. M., et al. (2002). Anovel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of B. Subtillis. J. Bacteriol. 184 5661–5671. 10.1128/JB.184.20.5661-5671.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entzminger K. C., Chang C., Myhre R. O., McCallum K. C., Maynard J. A. (2012). The Skp chaperone helps fold soluble proteins in vitro by inhibiting aggregation. Biochemistry 51 4822–4834. 10.1021/bi300412y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouet A. (2009). The surface of Bacillus anthracis. Mol. Aspect Med. 30 374–385. 10.1016/j.mam.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Freudl R. (2018). Signal peptides for recombinant protein secretion in bacterial expression systems. Microb. Cell Fact. 17:52. 10.1186/s12934-018-0901-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaza P., Osipiuk J., Wenta T., Zurawa-Janicka D., Jarzab M., Lesner A., et al. (2015). Structural and functional analysis of human HtrA3 protease and its subdomains. PLoS One 10:e0131142. 10.1371/journal.pone.0131142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen G., Hilgenfeld R. (2013). Architecture and regulation of HtrA-family proteins involved in protein quality control and stress response. Cell. Mol. Life. Sci. 70 761–775. 10.1007/s00018-012-1076-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey D. M., Browne P. J., Iavarone A. T., Teyra J., Lee E. H., Sidhu S. S., et al. (2016). Magnetite biomineralization in Magnetospirillum magmeticum is regulated by a switch-like behaviour in the HtrA protease MamE. J. Biol. Chem. 291 17941–17952. 10.1074/jbc.M116.731000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy B., Geppert T., Boehm M., Reisen F., Plattner P., Gadermayer G., et al. (2012). Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor supressor E-cadherin. J. Biol. Chem. 287 10115–10120. 10.1074/jbc.C111.333419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy B., Löwer M., Weydig C., Carra G., Tegtmeyer N., Geppert T., et al. (2010). Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11 798–804. 10.1038/embor.2010.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyrylainen H. L., Bolhuis A., Darmon E., Muukonen L., Koski P., Vitikainen M., et al. (2001). A novel two-component regulatory system in Bacillus subtillis for the survival of severe secretion stress. Mol. Microbiol. 41 1159–1172. 10.1046/j.1365-2958.2001.02576.x [DOI] [PubMed] [Google Scholar]
- Israeli M., Rotem S., Elia U., Bar-Haim E., Cohen O., Chitlaru T. (2016). A simple luminescent adenylate-cyclase functional assay for evaluation of Bacillus anthracis edema factor activity. Toxins 8:243. 10.3390/toxins8080243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanczyk J., Damjanovic D., Kooistra J., Leong V., Jomaa A., Ghirlando R., et al. (2007). Role of the PDZ domains in E. coli DegP protein. J. Bact. 189 3176–3186. 10.1021/bi701929m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Kim K. K. (2005). Structure and function of HtrA family proteins, the key players in protein quality control. J. Biochem. Mol. Biol. 38 266–274. 10.5483/BMBRep.2005.38.3.266 [DOI] [PubMed] [Google Scholar]
- Klimpel K. R., Arora N., Leppla S. H. (1994). Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol. Microbiol. 13 1093–1100. 10.1111/j.1365-2958.1994.tb00500.x [DOI] [PubMed] [Google Scholar]
- Koehler T. M. (2009). Bacillus anthracis physiology and genetics. Mol. Aspects Med. 30 386–396. 10.1016/j.mam.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacy T. M., Collier R. J. (2002). Structure and function of anthrax toxin. Curr. Top. Microbiol. Immun. 271 62–85. 10.1007/978-3-662-05767-4_4 [DOI] [PubMed] [Google Scholar]
- Leppla S. H. (1982). Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 79 3162–3166. 10.1073/pnas.79.10.3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy H., Weiss S., Altboum Z., Schlomovitz J., Glinert I., Sittner A., et al. (2012). Differential contribution of Bacillus anthracis toxins to pathogenicity in two animal models. Infect. Immun. 80 2623–2631. 10.1128/IAI.00244-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C., Skovierova H., Rowley G., Rezuchova B., Homerova D., Stevenson A., et al. (2009). Salmomella enterica Serovar Typhimurium HtrA: regulation of expression and role of the chaperone and protease activities during infection. Microbiology 155 873–881. 10.1099/mic.0.023754-0 [DOI] [PubMed] [Google Scholar]
- Lyon W. R., Caparon M. G. (2004). Role of the serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the Hemolysin Streptolysin S. Infect. Immun. 72 1618–1625. 10.1128/IAI.72.3.1618-1625.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MohamedMohaideen N. N., Palaninathan S. K., Morin P., Williams B. J., Braunstein M., Tichy S. E., et al. (2008). Structure and function of the virulence-associated high-temperature requirement A of Mycobacterium tuberculosis. Biochemistry 47 6092–6102. 10.1021/bi701929m [DOI] [PubMed] [Google Scholar]
- Noone D., Howell A., Collery R., Devine K. M. (2001). YkdA and YvtA, HtrA-like serine proteases in Bacillus subtillis engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183 654–663. 10.1128/JB.183.2.654-663.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Wren B. W. (1997). Th HtrA family of serine proteases. Mol. Microbiol. 26 209–221. 10.1046/j.1365-2958.1997.5601928.x [DOI] [PubMed] [Google Scholar]
- Pedersen L., Radulic M., Doric M., Abu Kwaik Y. (2001). HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Inf. Immun. 69 2569–2579. 10.1128/IAI.69.4.2569-2579.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantsev A. P., McCall R. M., Chahoud M., Hepler N. K., Fattah R., Leppla S. H. (2017). Genome engineering in Bacillus anthracis using tyrosine site-specific recombinases. PLoS One 12:e0183346. 10.1371/journal.pone.0183346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantsev A. P., Pomerantseva O. M., Moayeri M., Fattah R., Tallant C., Leppla S. H. (2011). A Bacillus anthracis strain deleted for six proteases serves as an effective host for production of recombinant proteins. Protein Expr. Purif. 80 80–90. 10.1016/j.pep.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela-Abramovich S., Chitlaru T., Gat O., Grosfeld H., Cohen O., Shafferman A. (2009). Novel and unique diagnostic biomarkers for Bacillus anthracis infection. Appl. Environ. Microbiol. 75 6157–6167. 10.1128/AEM.00766-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafferman A., Gat O., Ariel N., Chitlaru T., Grosfeld H., Zvi A., et al. (2010). “Reverse Vaccinology in B. anthracis,” in The Challenge of Highly Pathogenic Microorganisms, Mechanisms of Virulence and Novel Medical Countermeasures eds Shafferman A., Velan B., Ordentlich A. (Netherland: Springer Press; ) 295–306. [Google Scholar]
- Singh N., Kuppili R. R., Bose K. (2011). The structural basis of mode of activation and functional diversity: a case study with HtrA family of serine proteases. Arch. Biochem. Biophys. 516 85–96. 10.1016/j.abb.2011.10.007 [DOI] [PubMed] [Google Scholar]
- Skórko-Glonek J., Laskowska E., Sobiecka-Szkatula A., Lipinska B. (2007). Characterization of the chaperone-like activity of HtrA (DegP) protein from E. coli under the conditions of heat shock. Arch. Biochem. Biophys. 464 80–89. 10.1016/j.abb.2007.04.006 [DOI] [PubMed] [Google Scholar]
- Skórko-Glonek J., Wawrzynow A., Krzewski K., Kurpierz K., Lipinska B. (1995). Site-directed mutagenesis of the HtrA(DegP) serine protease, whose proteolytic activity is indispensable for E. coli survival at elevated temperatures. Gene 163 47–52. 10.1016/0378-1119(95)00406-V [DOI] [PubMed] [Google Scholar]
- Skórko-Glonek J., Zurawa D., Tanfani F., Scire A., Wawrzynow A., Narkiewicz J., et al. (2003). The N-terminal region of HtrA heat shock protease from E. coli is essential for stabilization of HtrA primary structure and maintaining of its oligomeric structure. Biochem. Byophis. Acta 1649 171–182. 10.1016/S1570-9639(03)00170-5 [DOI] [PubMed] [Google Scholar]
- Skórko-Glonek J., Zurawa-Janicka D., Koper T., Jarzab M., Figaj D., Glaza P., et al. (2013). HtrA protease family as therapeutic targets. Curr. Pharm. Des. 19 977–1009. 10.2174/1381612811319060003 [DOI] [PubMed] [Google Scholar]
- Spiess C., Beil A., Ehrmann M. (1999). A temperature dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97 339–347. 10.1016/S0092-8674(00)80743-6 [DOI] [PubMed] [Google Scholar]
- Tegtmeyer N., Moodley Y., Yamaoka Y., Traverso F. R., Pernitzch S. R., Schmidt V., et al. (2016). Characterization of worldwide Helicobacter pylori strains reveals genetic conservation and essentiality of serine protease HtrA. Mol. Microbiol. 99 925–944. 10.1111/mmi.13276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H., Antelman H., Jongbloed J. D. H., Braun P. G., Darmon E., Dorenbos R., et al. (2004). Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Molec. Biol. Rev. 68 207–233. 10.1128/MMBR.68.2.207-233.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truebestein L., Tennstaedt, Monig T., Krojer T., Canellas F., Kaiser M.et al. (2011). Substrate-induced remodeling of the active site regulates human HTRA1 activity. Nat. Struct. Mol. Biol. 18 386–388. 10.1038/nsmb.2013 [DOI] [PubMed] [Google Scholar]
- Tsirigotaki A., De Geiter J., Šoštarić N., Economou A., Karamanou S. (2017). Protein export through the bacterial Sec pathway. Nat. Rev. Microbiol. 15 21–36. 10.1038/nrmicro.2016.161 [DOI] [PubMed] [Google Scholar]
- Turnbull P. C. B. (1991). Anthrax vaccines: past, present and future. Vaccine 9 533–539. 10.1016/0264-410X(91)90237-Z [DOI] [PubMed] [Google Scholar]
- Vitikainen M., Hyyrylainen H.-L., Kivimaki A., Sarvas M. (2005). Secretion of heterologous proteins in Bacillus subtillis can be improved by engineering cell components affecting post-translocational protein folding and degradation. J. Appl. Microbiol. 99 363–375. 10.1111/j.1365-2672.2005.02572.x [DOI] [PubMed] [Google Scholar]
- Walsh N. P., Alba B. M., Bose B., Gross C. A., Sauer R. T. (2003). OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113 61–71. 10.1016/S0092-8674(03)00203-4 [DOI] [PubMed] [Google Scholar]
- Welkos S. L., Bozue J. A., Cote C. K. (2011). “The interaction between Bacillus anthracis and macrophages,” in Bacillus anthracis and anthrax ed. Bergman N. H. (Hoboken, NJ: John Wiley & Sons, Inc. Press; ) 179–208. [Google Scholar]
- Wilken C., Kitzing K., Kurzbauer R., Ehrmann M., Clausen T. (2004). Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117 483–494. 10.1016/S0092-8674(04)00454-4 [DOI] [PubMed] [Google Scholar]
- Zhu F., Yang X., Wu Y., Wang Y., Tang X.-F., Tang B. (2017). Release of an HtrA-like protease from the cell surface of thermophilic Brevibacillus sp. WF146 via substrate-induced autoprocessing of the N-terminal membrane anchor. Front. Microbiol. 8:481. 10.3389/fmicb.2017.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


