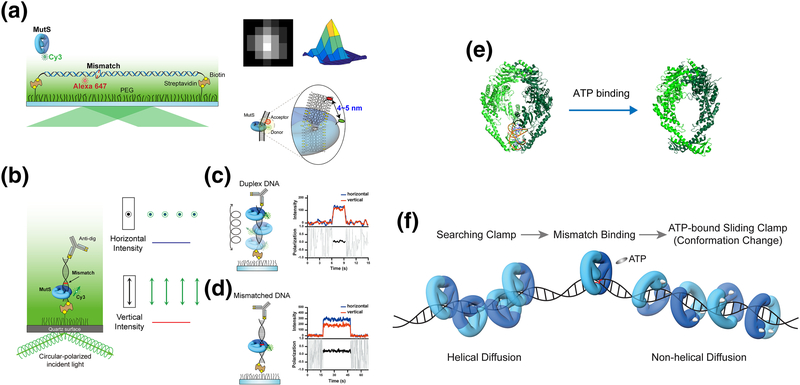
Figure 2. Conformational switch of MutS.
(a) Mismatched DNA containing a dG/dT mismatch or an unpaired thymine (dT) mismatch was stretched and immobilized on a flow-cell surface with defined hydrodynamic drag by applying a laminar flow. A single acceptor fluorophore was attached to the mismatched DNA to monitor the mismatch binding by MutS that was tagged with a single donor fluorophore using a single-molecule Förster Resonance Energy Transfer assay. The time-dependent position of MutS on the DNA was determined with an accuracy of a few nanometers using 2D Gaussian-fitted center of intensity profiles [104, 105]. (b) A schematic representation of the single-molecule fluorescence polarization system to measure the polarization of Cy3-MutS. (c) The emission signals of Cy3-MutS on duplex DNA was depolarized. (d) The emission signals of Cy3-MutS on mismatched DNA was strongly polarized, which indicates that the Cy3-MutS was trapped at the mismatch. (e) The crystal structures of E. coli MutS bound to a mismatch and ATP-bound MutS. (PDB: 1e3M [40], mismatch-bound MutS and 5akc [47], ATP-bound MutS). (f) TaMutS forms two different sliding clamps on mismatched DNA: a mismatch searching MutS clamp (~1 s) that diffuses along the DNA in a continuous contact with the DNA backbone (helical diffusion) and a highly stable (~600 s) ATP-bound sliding clamp in a discontinuous contact with the DNA backbone (non-helical diffusion).