Abstract
Aims
The JAK2 V617F mutation is highly recurrent in many of the myeloproliferative neoplasms, a molecular variant that can be easily detected using sensitive and minimally invasive techniques. Given the ease of JAK2 V617F testing, this test may be improperly requested for the purposes of patient ‘screening’ and to optimise laboratory resource utilisation, it behooves clinicians and laboratorians to perform JAK2 V617F testing only when most appropriate.
Methods
To assist with the screening of patients being considered for JAK2 V617F testing, we developed a clinical decision rule, “JAK2-tree”, which can be easily applied to basic CBC parameters (haemoglobin, platelet and white blood cell counts).
Results
We tested JAK2-tree on two independent datasets, one an unselected population-based sample (the Copenhagen General Population Study) and the other an historical clinical laboratory referral set, with sensitivities for JAK2 V617F detection of 91% and 94%, respectively. As applied to the historical laboratory referral dataset, moreover, the JAK2-tree algorithm would have reduced JAK2 V617F testing volume over the period of evaluation by 15%.
Conclusions
Our work supports a simple decision-tree-based screening approach to optimize the selection of patients most appropriate for JAK2 V617F testing.
Keywords: myeloproliferative disease, clinical audit, laboratory tests, molecular pathology, laboratory management
Background
The myeloproliferative neoplasms (MPNs) are relatively common, with recent data suggesting an incidence of 11.8 per 100,000.1 Rather than representing a single disease entity, the MPNs form a spectrum of clinically and biologically variable diseases.2 Despite this heterogeneity, however, the MPNs share the commonality of frequent Janus Kinase 2 (JAK2) gene mutations when chronic myeloid, chronic neutrophilic and chronic eosinophilic leukaemias are excluded.2
The JAK2 gene, mapped to locus 9p24, encodes a tyrosine kinase involved in cytokine receptor signalling through the STAT pathway.3 4 When constitutively activated, most often by way of the JAK2 p.Val617Phe (V617F) mutation, the result is an over-responsive kinase to even low-level cytokine stimulation (including erythropoietin).5 The JAK2 V617F mutation has been identified with variable frequency across the MPNs; in approximately 96% of polycythemia vera (PV) cases, 55% of essential thrombocytosis (ET) cases and in approximately 65% of primary myelofibrosis (PMF) cases.6 The JAK2 V617F mutation has therefore become an important part of the diagnostic criteria for these neoplasms.7
Despite the high frequency of JAK2 mutations in diagnosed MPNs, and hence the seeming appeal of JAK2 mutation testing as a screening tool, there are a number of caveats to JAK2 testing that narrow its applicability. While the JAK2 V617F mutation is most common, other JAK2 mutations may be seen in a minority of MPNs, such as mutations in exon 12 detected in ~3%–4% of patients with PV.6 8 These observations highlight the imperfect sensitivity of testing only for the JAK2 V617F mutation. Indeed, a population-level screening study for the JAK2 V617F mutation suggested a very low sensitivity of only 23% testing for the subsequent diagnosis of an MPN.9 Instead, the actual diagnostic value of JAK2 V617F mutation testing relates to its noted specificity approaching 100% for the diagnosis of an MPN.9 Also worthy of consideration is the variety of testing methods available, ranging from quantitative PCR (qPCR), to allele specific PCR, and Sanger sequencing, each with its attendant limits of analytical sensitivity and (usually inversely related) associated cost.6 Given the wide range of potential JAK2 V617F mutant allele fractions that may be encountered in MPNs (with levels as low as 1%), laboratories are encouraged to offer JAK2 V617F mutation testing at high analytical sensitivity (typically requiring qPCR-based methods).6 The high analytical sensitivity of current JAK2 V617F mutation testing methods makes peripheral blood detection possible, rather than requiring more invasive testing of clonally enriched lesional tissues. The consequences of easily requested non-invasive testing, while attractive to patients and clinicians alike, might include misuse as a pseudo-screening procedure.
Our laboratory network has been performing JAK2 V617F mutation testing for nearly two decades. Since the inception of our testing protocol (employing a sensitive qPCR method), we have noted a progressive increase in the volume of requested testing, countered by a progressive decrease in the hit rate of our assay (ie, the fraction of positive cases among the total tested), despite stable quality metrics. These anecdotal observations support the hypothesis of systematic misuse. For these reasons, we set to ascertain whether a carefully crafted algorithm, incorporating simple peripheral blood parameter assessment using a decision-tree approach, might permit our laboratory (and our clinical colleagues alike) to better ‘screen’ JAK2 V617F mutation testing requests for clinical appropriateness.
Methods
All aspects of this project conformed to the tenets of the Declaration of Helsinki. Data collection and processing through Calgary Lab Services conformed to the Alberta Research Ethics Community Consensus Initiative guidelines for the undertaking of Quality Assurance Projects.10 Statistical computations were performed in either Excel (V.15.41, Microsoft, Seattle, Washington, USA) or MATLAB (V.R2016b, The MathWorks, Natick, Massachusetts, USA); in general, p values <0.05 were considered statistically significant.
JAK2-tree design
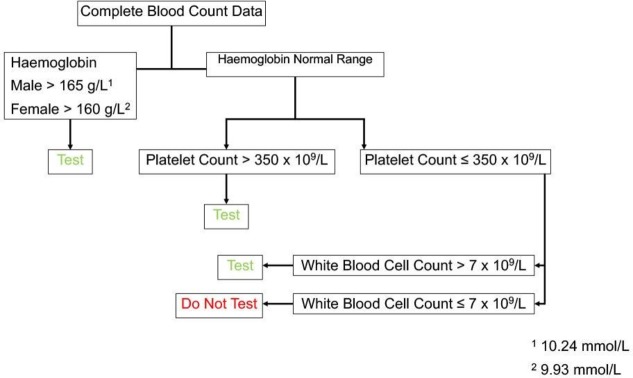
We employed the rubric of the WHO Classification of Tumours of the Haematopoietic and Lymphoid Tissues2 as the basis for our CBC Parameter Decision-Tree Algorithm (JAK2-tree) design. Beginning with consideration of the utility of JAK2 V617F mutation testing in the setting of PV, we set the first branch point of our JAK2-tree to be a haemoglobin (Hb) cut-off of (greater than) 165 g/L in men or 160 g/L in women. (The other erythroid parameters that are included in the WHO rubric for PV diagnosis, namely haematocrit and red cell mass, were not incorporated in JAK2-tree owing to the fact that the former is typically a calculated parameter and the latter is not routinely measured). The second branch point (in cases not exceeding the cut-offs of the first branch point) was set to address the possibility of thrombocytosis, as might be seen in the context of ET, PV and PMF.11 At this point, rather than set the platelet cut-off count equal to that of the WHO rubric, we opted to use a more conservative cut-off of (greater than) 350×109/L, in line with recent data suggesting that a lower high-platelet cut-off might be more appropriate, especially in older populations.12 The final JAK2-tree branch point (in cases not exceeding the cut-off of the second branch point) was set to address the possibility of abnormal white-blood cell counts (WBC), which might factor into the WHO rubric for the diagnosis of PMF or pre-PMF. We again applied a highly conservative cut-off of WBC (greater than) 7×109/L in order to exclude only those cases with low WBC (noting that some MPNs may have normal or near-normal WBC).13 14 The JAK2-tree is presented graphically in figure 1.
Figure 1.
JAK2-tree algorithm.
JAK2-tree validation using population data
The Copenhagen General Population Study, an ongoing population-based prospective cohort study initiated in 2003, served to provide the most robust estimate of JAK2 V617F mutation prevalence to date,9 and by virtue of accompanying CBC data, has served to support the diagnostic value of JAK2 testing. Within this dataset, 50 363 subjects from the general population had peripheral blood samples collected, from which CBC data were obtained, as well as JAK2 V617F allele ratio testing by qPCR-based methods as previously described.9 Within this dataset, a total of 68 JAK2 V617F positive cases were detected at an allele ratio of at least 0.8%. The JAK2-tree algorithm was applied to the Copenhagen General Population Study to assess the utility of the decision tree in an unselected population-based sample. JAK2-tree algorithm performance analyses also included data on age and sex.
JAK2-tree validation using historical laboratory testing data
We reviewed the electronic records of all JAK2 V617F mutation tests performed through Calgary Lab Services, beginning in January 2009 (when fully electronic JAK2 V617F mutation reporting became available in our health region) through to the end of August 2017 (when the data review was initiated). In concert, we obtained the appertaining CBC data derived from all patient encounters within the prior year. With these data were also included age and sex. It bears noting that, in the vast majority of cases, ‘clinical history’ or ‘indications for testing’ were not provided. The collected data were input into an electronic version of the JAK2-tree (implemented in MATLAB, V.R2016b, The MathWorks) to compute metrics of JAK2-tree predictive accuracy for JAK2 V617F mutation positivity.
Results
The Copenhagen General Population Study
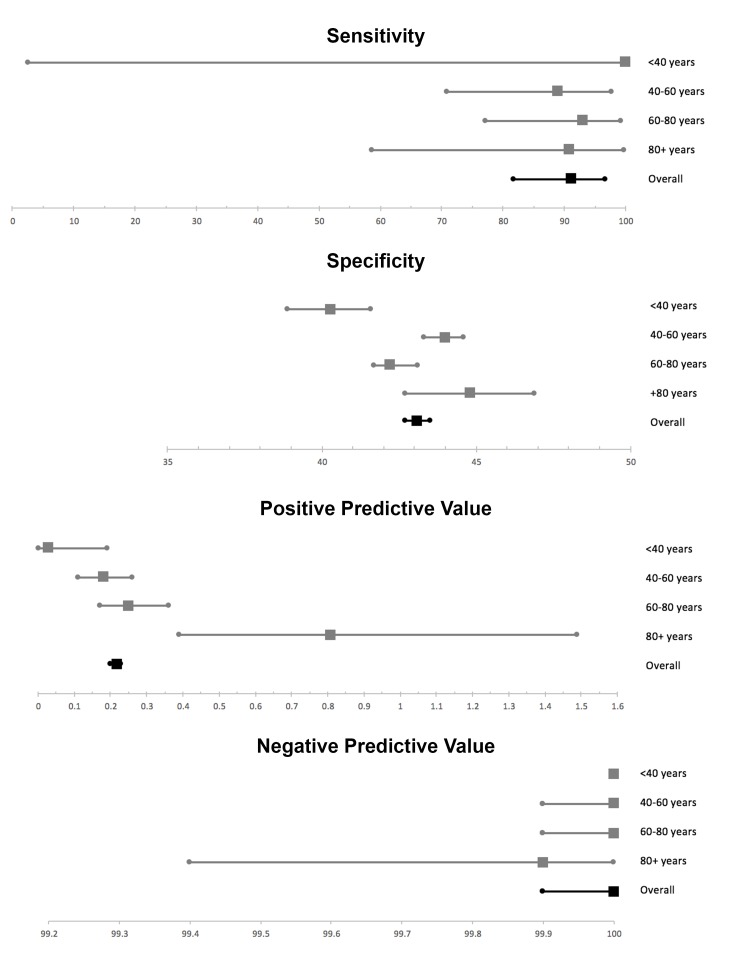
Among the included 50 363 individuals, the mean age was 57 years (range 20–99 years) and 45% were female. Table 1 outlines the confusion matrix of the JAK2-tree algorithm applied to the Copenhagen Population Study. The JAK2-tree algorithm demonstrated a sensitivity of 91% (95% CI 82% to 97%) and a negative predictive value of 99% (95% CI 99% to 100%). Not surprisingly, given its purposes as a screening tool, JAK2-tree yielded a low specificity of 43% (95% CI 42% to 44%) and a positive predictive value of 0.22% (95% CI 0.20% to 0.23%) when applied to the Copenhagen General Population Study. Table 2 highlights the characteristics of the six JAK2 V617F-positive cases ‘missed’ by the JAK2-tree algorithm. From among these six ‘missed’ cases, only three had an appertaining malignant diagnosis: one had an already prevalent diagnosis of MPN, one was concomitantly diagnosed with an MPN and the third developed acute myeloid leukaemia during follow-up. Figure 2 highlights the performance characteristics of the JAK2-tree algorithm broken down by age bracket.
Table 1.
Copenhagen General Population Study JAK2-tree confusion matrix
| JAK2 V617F status (%) | ||
| Positive | Negative | |
| JAK2-tree algorithm (%) | ||
| Test | 62 (0.12%) | 28 623 (57%) |
| Do not test | 6 (0.01%) | 21 672 (43%) |
Table 2.
Characteristics at blood sampling of the six cases from the Copenhagen General Population Study ‘missed’ by the JAK2-tree algorithm.
| Age, years | Sex, per cent women | JAK2 V617F (allele burden %) | Haemoglobin (g/L) | Platelet count (x 109/L) | Leucocyte Count (x 109/L) | Haematological diagnoses* |
| 60 (14) | 50 | 5.4 (3.9) | 140 (11) | 232 (78) | 5.5 (1.2) | One incident case of acute myeloid leukaemia, pmeone prevalent and one incident case of myeloproliferative neoplasm |
Values are per cent for sex and mean (SD) for continuous variables.
*Follow-up until 31 December 2016.
Figure 2.
Performance characteristics of the JAK2-tree algorithm, as applied to the Copenhagen General Population Study, broken down by age bracket.
Historical laboratory testing data
We reviewed the records of all JAK2 V617F mutation tests performed through Calgary Lab Services from January 2009 through to the end of August 2017, amounting to 2796 days, yielding a total of 2989 JAK2 V617F mutation test records, of which 580 JAK2 V617F mutation tests were reported positive (yielding an overall ‘hit-rate’ of 19%). Seventy-four per cent of JAK2 V617F mutation test specimens were collected on the same day as a CBC specimen, and 95% of JAK2 V617F mutation test specimens had CBC data available from within the 44 preceding days (range 0 to –364 days).
We tallied 2916 unique patient identifiers from among the 2989 JAK2 V617F mutation test records. Fifty-two per cent of patients evaluated were female, and the mean overall age was 58 years (range 3–98). Of interest, 64 patients (2%) were tested more than once. Of these repeat test cases, 60% were performed within the same year. Also, of the 64 patients tested more than once, none were found later positive after an initial negative result and 23% consisted of repeat tests of previously positive cases.
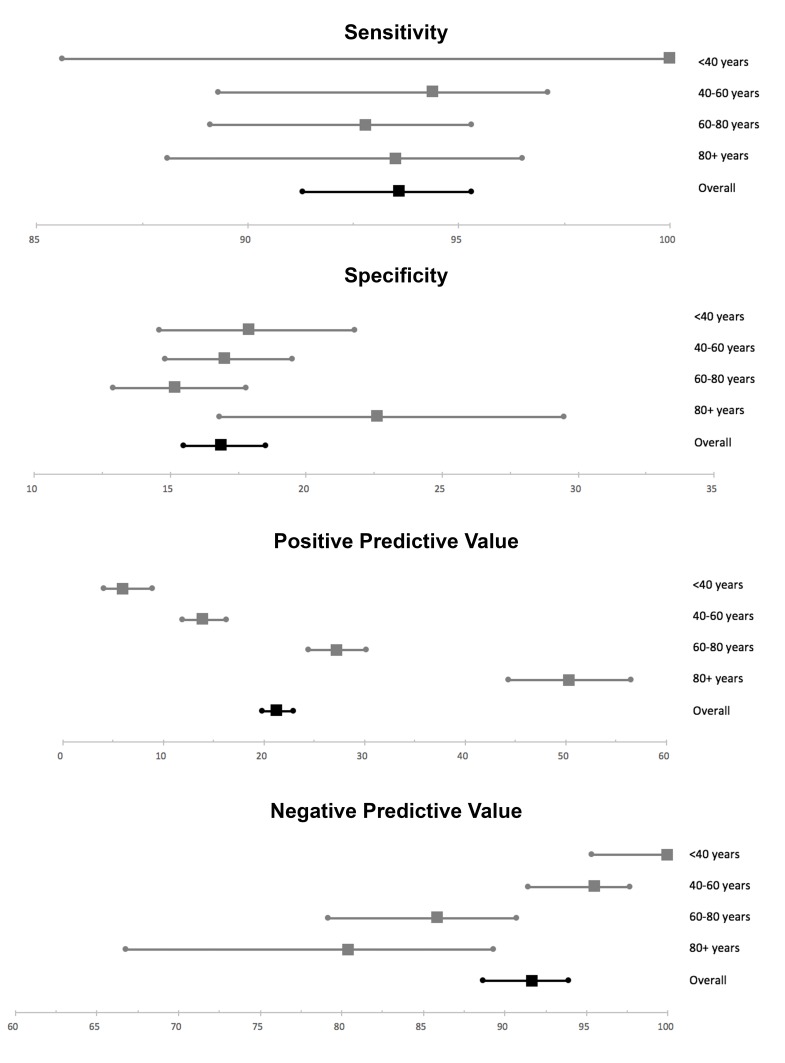
Table 3 outlines the JAK2-tree algorithm results applied to full cohort, and figure 3 highlights the performance characteristics of the JAK2-tree algorithm broken down by age bracket. The algorithm demonstrated a sensitivity of 94% (95% CI 91% to 95%) for prediction of JAK2 V617F mutation, and a negative predictive value of 92% (95% CI 89% to 94%). Not surprisingly, given its purpose as a screening tool, JAK2-tree was noted to have a low specificity of only 17% (95% CI 16% to 19%) and a low positive predictive value of only 21% (95% CI 20% to 23%). Of additional interest, however, is that JAK2-tree would have missed only 1.2% of JAK2 V617F mutation-positive cases (the false-negative estimate), while reducing the number of tests performed by 15%. Additionally, applying the JAK2-tree algorithm, and excluding those cases ‘screened negative’, the estimated hit-rate of JAK2 V617F mutation detection would have been approximately 21%, a number insignificantly different from the actual hit rate of 19% (p=0.073).
Table 3.
Calgary Lab Services retrospective dataset JAK2-tree confusion matrix
| JAK2 V617F status (%) | ||
| Positive | Negative | |
| JAK2-tree algorithm (%) | ||
| Test | 543 (18%) | 2001 (67%) |
| Do not test | 37 (1.2%) | 408 (14%) |
Figure 3.
Performance characteristics of the JAK2-tree algorithm, as applied to the Calgary Lab Services historical JAK2 V617F clinical testing dataset, broken down by age bracket.
Given the availability of a number of patients with multiple CBCs performed preceding JAK2 testing, we tested tandem application of the JAK2-tree algorithm. For this, we applied the JAK2-tree algorithm to all patients for whom more than one CBC data point was available, separated by at least 3 months; if the JAK2-tree algorithm on first CBC testing did not indicate testing, the next available CBC data (obtained 3 months subsequent) were assessed to see if JAK2 V617F testing was indicated by the JAK2-tree algorithm. Using this tandem approach, there was no significant difference in algorithm performance (F-statistic=0.18; p=0.68).
Discussion
We propose a simple CBC-based screening tool for laboratory use, aimed to identify cases most appropriate for JAK2 V617F mutation testing. Such tools are strongly warranted owing to increased demands for molecular testing. As noted by careful examination of our own historical JAK2 V617F test dataset, such tools might also be indicated to counter laboratory test ‘abuse’.
The JAK2-tree algorithm can be easily applied using the three most widely available CBC parameters (Hb, platelet count and WBC). The JAK2-tree approach could be easily incorporated into the background logic of electronic laboratory accessioning/records systems to flag inappropriate testing requests. When applied to an historical laboratory referral dataset, the JAK2-tree algorithm demonstrated a sensitivity of 94% and negative predictive value of 92% for the prediction of a JAK2 V617F mutation. Noting the inherently biased nature of this dataset, we also evaluated the algorithm over the most robust population-based JAK2 V617F testing dataset available (the Copenhagen General Population Study), in which comparable results were noted (sensitivity of 91% and negative predictive value of 99%). From our historical dataset, the JAK2-tree approach would have reduced JAK2 V617F requests by 15%, with a false-negative error estimate of only 1.2%.
As with all clinical decision tools, care should be taken when using the JAK2-tree approach, owing to certain limitations. In the Copenhagen General Population Study, the JAK2 V617F mutation was detected by a PCR-based TaqMan screening assay identifying the 1% with the highest signal, these were subsequently tested with a highly sensitive real-time qPCR assay with a limit of detection of 0.8%.9 Patients with MPNs have been reported to have allele burdens as low as 0.01%–0.34%, depending on assay type at diagnosis.15 Thus, individuals with low burdens of JAK2 V617F were not identified in the Copenhagen General Population Study. Therefore, despite the promising results in the present study, the need for further validation of the JAK2-tree in other general population cohorts with measurement of the JAK2 V617F mutation burden using more sensitive assays such as ddPCR-based assays would be warranted. Moreover, the individuals from the Copenhagen General Population Study were all white and of Danish descent; therefore, these results may not necessarily apply to other ethnicities.
Importantly, the JAK2-tree was not designed to be used in place of clinical acumen, and other specific clinical indications for testing that might be brought to bear in specific circumstances. Indeed, there may be specific and highly significant examples of non-CBC informed clinical indications that warrant JAK2 V617F testing. Such examples might include thrombotic events involving atypical sites16; to provide molecular confirmation of a previous clinical diagnosis of an MPN; or for patients in which JAK2 V617F allele burden determination might influence therapy.17 In such instances, available CBC data might not support testing, clinical information notwithstanding. As such, laboratories are cautioned to defer to specific clinical history indicators and careful laboratory-to-clinic communication in this regard, as required. The JAK2-tree algorithm also does not distinguish between leukocytosis owing to lymphocytes or myeloid forms; in the context of isolated lymphocytosis, and without other clinical indications, JAK2 V617F testing would likely be unremarkable, despite algorithmic suggestion to the contrary.
Although we could not demonstrate significant changes in performance parameters with tandem application of the JAK2-tree algorithm, other algorithmic variations might be worth testing. For instance, assuming normal Hb and platelet count, the JAK2-tree approach suggests that a normal WBC would be sufficient for testing. Instead, and considering the minor WHO criteria for diagnosis of PMF, a laboratory might adjust the JAK2-tree algorithm to stipulate reflexive testing with an elevated WBC, and with a signal to discuss the request with the clinical team in instances of normal range (ie, 7–11×109/L) WBC to confirm appropriateness.
Take home messages.
Laboratory testing, especially testing employing molecular methods, should be appropriately optimised for the clinical scenario.
Clinical decision rules applied to laboratory testing can reduce costs and prevent waste.
Simple clinical decision rules like JAK2-tree can assist in appropriate test ordering.
Acknowledgments
The authors wish to thank the Calgary Lab Services Data Management Teams, as well as the members of the Molecular Haematology Laboratory, for their contributions to this project. They are indebted and thankful to all participants and staff from the Copenhagen General Population Study for their valuable contributions.
Footnotes
Handling editor: Prof Mary Frances McMullin.
Contributors: This project was conceived by MTSR, FK, GS and EM, with CLS data procurement facilitated by TL. The Copenhagen Population Study was led by KMP, YÇ and SEB. Current data analysis was facilitated by KMP. The manuscript was largely written by EM with the assistance of KMP. All authors reviewed and approved of the manuscript.
Competing interests: None declared.
Patient consent: Not required.
Ethics approval: The Copenhagen General Population Study was approved by the Herlev and Gentofte Hospital and by a Danish ethical committee (approval number: H-KF-01-144/01).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. National Cancer Institute SRP. SEER*Explorer: An interactive website for SEER cancer statistics. Available: https://seer.cancer.gov/explorer/
- 2. Weltgesundheitsorganisation WHO classification of tumours of haematopoietic and lymphoid tissues : Swerdlow SH, Campo E, Harris NL, World Health Organization classification of tumours. 4th edn Lyon: International Agency for Research on Cancer, 2017: 585. [Google Scholar]
- 3. Dalal I, Arpaia E, Dadi H, et al. . Cloning and characterization of the human homolog of mouse Jak2. Blood 1998;91:844–51. [PubMed] [Google Scholar]
- 4. Schwaller J, Parganas E, Wang D, et al. . Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell 2000;6:693–704. 10.1016/S1097-2765(00)00067-8 [DOI] [PubMed] [Google Scholar]
- 5. James C, Ugo V, Le Couédic JP, et al. . A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434:1144–8. 10.1038/nature03546 [DOI] [PubMed] [Google Scholar]
- 6. Gong JZ, Cook JR, Greiner TC, et al. . Laboratory practice guidelines for detecting and reporting JAK2 and MPL mutations in myeloproliferative neoplasms: a report of the Association for Molecular Pathology. J Mol Diagn 2013;15:733–44. 10.1016/j.jmoldx.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 7. Barbui T, Thiele J, Gisslinger H, et al. . The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: document summary and in-depth discussion. Blood Cancer J 2018;8:15 10.1038/s41408-018-0054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Passamonti F, Rumi E, Pietra D, et al. . A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia 2010;24:1574–9. 10.1038/leu.2010.148 [DOI] [PubMed] [Google Scholar]
- 9. Nielsen C, Birgens HS, Nordestgaard BG, et al. . Diagnostic value of JAK2 V617F somatic mutation for myeloproliferative cancer in 49 488 individuals from the general population. Br J Haematol 2013;160:70–9. 10.1111/bjh.12099 [DOI] [PubMed] [Google Scholar]
- 10.Alberta Research Ethics Community Consensus Initiative (ARECCI) Network. ARECCI ethics screening tool. 2005. Available: http://www.aihealthsolutions.ca/arecci/screening/240345/529e8675083049bf925aa8d88140507c
- 11. Spivak JL. Myeloproliferative Neoplasms. N Engl J Med 2017;376:2168–81. 10.1056/NEJMra1406186 [DOI] [PubMed] [Google Scholar]
- 12. Bonaccio M, Di Castelnuovo A, Costanzo S, et al. . Age-sex-specific ranges of platelet count and all-cause mortality: prospective findings from the MOLI-SANI study. Blood 2016;127:1614–6. 10.1182/blood-2016-01-692814 [DOI] [PubMed] [Google Scholar]
- 13. Barosi G, Rosti V, Bonetti E, et al. . Evidence that prefibrotic myelofibrosis is aligned along a clinical and biological continuum featuring primary myelofibrosis. PLoS One 2012;7:e35631 10.1371/journal.pone.0035631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buxhofer-Ausch V, Gisslinger B, Schalling M, et al. . Impact of white blood cell counts at diagnosis and during follow-up in patients with essential thrombocythaemia and prefibrotic primary myelofibrosis. Br J Haematol 2017;179:166–9. 10.1111/bjh.14202 [DOI] [PubMed] [Google Scholar]
- 15. Link-Lenczowska D, Pallisgaard N, Cordua S, et al. . A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann Hematol 2018;97:2299–308. 10.1007/s00277-018-3451-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shetty S, Kulkarni B, Pai N, et al. . JAK2 mutations across a spectrum of venous thrombosis cases. Am J Clin Pathol 2010;134:82–5. 10.1309/AJCP7VO4HAIZYATP [DOI] [PubMed] [Google Scholar]
- 17. Vannucchi AM, Pieri L, Guglielmelli P. JAK2 allele burden in the myeloproliferative neoplasms: effects on phenotype, prognosis and change with treatment. Ther Adv Hematol 2011;2:21–32. 10.1177/2040620710394474 [DOI] [PMC free article] [PubMed] [Google Scholar]