Abstract
Background
The C. macropomum is a characiform fish from the Amazon basin that has been hybridized with other pacu species to produce commercial hybrids, such as the tambacu. However, little is known of the functional genomics of the parental species or these hybrid forms. The transcriptome of C. macropomum and tambacu were sequenced using 454 Roche platform (pyrosequencing) techniques to characterize the domains of Gene Ontology (GO) and to evaluate the levels of gene expression in the two organisms.
Results
The 8,188,945 reads were assembled into 400,845 contigs. A total of 58,322 contigs were annotated with a predominance of biological processes for both organisms, as determined by Gene Ontology (GO). Similar numbers of metabolic pathways were identified in both the C. macropomum and the tambacu, with the metabolism category presenting the largest number of transcripts. The BUSCO analysis indicated that our assembly was more than 40% complete. We identified 21,986 genes for the two fishes. The P and Log2FC values indicated significant differences in the levels of gene expression, with a total of 600 up-regulated genes.
Conclusion
In spite of the lack of a reference genome, the functional annotation was successful, and confirmed a considerable difference in the specificity and levels of gene expression between the two organisms. This report provides a comprehensive baseline for the genetic management of these commercially important fishes, in particular for the identification of specific genes that may represent markers involved in the immunity, growth, and fertility of these organisms, with potential practical applications in aquaculture management.
Background
Next Generation Sequencing (NGS) technologies have provided a range of potential markers for both model and non-model species through the discovery of new genes from an enormous database [1–3]. Over the past few years, NGS has been used increasingly to characterize the transcripts of a number of different species and tissues, in addition to contributing to the evaluation of gene expression in commercial lineages. Fish are especially important, given that they comprise a highly diversified group, with many economically important species, but little information on their functional genomics. The FLX (GS FLX) Genome sequencing system, which uses the 454 sequencing approach, is one of the most widely-used and successful approaches to the evaluation of the fish transcriptome [4–6].
The tambaqui (Colossoma macropomum) is an important fishery resource in the Amazon region, and is distributed naturally in both the Amazon and Orinoco river basins. Given its aquacultural performance, due to its rapid growth and robustness, the C. macropomum is currently the most common species raised on Brazilian fish farms [7, 8], and is becoming increasingly popular in other tropical countries. As the second largest South American freshwater fish, reaching 90 cm and 30 kg, the C. macropomum is especially suitable for tropical aquaculture, especially as it is also extremely fertile and robust, and can adapt easily to polyculture systems [7]. This species has also often been hybridized with other pacus, in particular Piaractus mesopotamicus [9, 10], producing a hybrid known as the tambacu. The tambacu results from the induced crossing of the female C. macropomum with the male P. mesopotamicus, and has been raised ever more intensively in recent years, replacing its parental species in tropical aquaculture operations [8].
The few comparative studies available for the C. macropomum and tambacu have focused on growth, genetics, biochemistry, and the immune system [11–13], while no comparative analyses of their functional genomics have yet been published. In fact, few data are available on the functional genomics of C. macropomum. However, Prado-Lima and Val [14] did analyze the transcriptome of the muscle of juvenile t C. macropomum, subjected to variation in temperature. In this experiment, significant differences were observed in the expression of specific groups of genes in responses to changes in the ambient conditions. Gomes et al. [15] characterized the miRNAs of the liver and muscle of juvenile C. macropomum, and identified a number of conserved miRNAs that were shared between the two types of tissue, albeit with distinct levels of expression. Studying 30 C. macropomum specimens, Martinez et al. [16] analyzed nuclear Single Nucleotide Polymorphisms (SNPs) using Next Generation Sequencing-based genotyping. Using the genotyping by sequencing method, Nunes et al. [17] identified a large number of SNPs in C. macropomum, and constructed a high-resolution genetic linkage map for a full-sibling family of 124 individuals, including their parents. Apart from these studies, there are no data on the transcriptome of the tambacu or any comparative analyses involving its ancestral species.
The transcriptome is the initial product of genome expression and is the principal focus of the investigation of gene function [18, 19]. Thus, we compared the transcriptome of C. macropomum and tambacu to characterize the Gene Ontology (GO) domains and metabolic pathways, as well as evaluating the levels of gene expression in the two organisms. Two types of tissue–skin and muscle–were chosen for these analyses. The skin represents the main defensive barrier of a vertebrate, and presents unique biological processes related to its complex structure and cell composition [20, 21]. Skeletal muscle represents about half of the body mass of a fish, and is associated with the swimming capabilities of these animals [22]. The results of this study are important for the conservation of the species, and the markers may also have practical applications for management of farmed stocks.
Material and methods
Ethics statement
The samples analyzed in the present study were obtained from fish farms and collected with the permission of the Brazilian federal environmental, authorized by the Chico Mendes Institute for Biodiversity (agency ICMBio) through license number 12.773–1 granted to Iracilda Sampaio. The animal experimentation conducted was in accordance with the National control Board of Animal Experimentation (CONCEA), Brazilian federal law no. 11.794, and the International guidelines defined by the U.K. Animals (Scientific Procedures) Act, 1986. The specimens were then euthanized by immersion in anesthetic Benzocaine Hydrochloride, as recommended by the Brazilian Federal Council of Veterinary Medicine (federal law 5,517, resolution CFMV no. 1000/2012). All efforts were made to minimize the intensity and duration of distress and exposure to pain.
Sample collection
The C. macropomum and tambacu specimens were collected from the Andrera fish farm in Bonito, in Pará state, Brazil. All specimens were identified prior to collection, based on morphological traits, such as the shape of the body, and the number of rakers on the first gill arch. However, given the considerable morphological similarities between the C. macropomum and its hybrids, genetic tests were conducted to confirm the identity of the specimens. As result, eight adults from C. macropomum and tambacu were selected, with both groups presenting a mean body length of 0.6 ± 0.2 m and weight of 3 ± 0.3 kg. The specimens were euthanized for the removal of fragments of the skin and muscle. The tissue samples were stored immediately in 1 mL of RNAlater (Ambion, Life Technologies, USA) at 4°C for 24 hours and then transferred to an ultrafreezer at -80°C. To ensure the identification of specimens (pure vs. hybrids), fragments of muscle were also stored in 100% ethanol at -20°C for DNA extraction.
It is important to note that the C. macropomum is a rheophilic species that is unable to breed in captivity [7]. These fish present obvious sexual dimorphism only during the breeding period, when the females acquire a soft, rounded abdomen, as well as a prominent, reddish urogenital papilla, while the male expel fluid semen after gentle abdominal massage [23]. As the specimens were collected during the non-breeding period, none of the individuals presented these characteristics, which impeded the differentiation of males and females.
DNA extraction, PCRs and sequencing
The total DNA was isolated using phenol-chloroform and precipitation in ethanol [24]. The integrity of the DNA samples was verified by electrophoresis in 1% agarose gel stained with GelRed, followed by visualization under UV light.
The isolation and amplification of the mitochondrial Control Region (D-loop) was based on Polymerase Chain Reaction (PCR), as described by Gomes et al. [13]. The products of the amplification were sequenced using the Big Dye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems), followed by electrophoresis in an automatic sequencer (Genetic Analyzer 3500). The sequences were then edited and aligned in BIOEDIT 7.2.5 [25]. The identification of the mitochondrial maternal lineage was based on comparisons with the sequences of the native species provided by Gomes et al. [13] and the sequences available in GenBank (Colossoma macropomum DQ480074 and Piaractus mesopotamicus AF283959).
Multiplex PCR
The C. macropomum and hybrids individuals were also identified by multiplex PCR using the nuclear α-tropomiosin, as described by Gomes et al. [13]. We, then, used the universal primer Tropo Serra F (5’-GAGTTGGATCGGGCTCAG-3’) and species-specific primers (Tropo Cm R 5’-ATACAACAATGCCATCGCT-3’ and Tropo Pm R 5’-CTTCAGCTGGATCTCCTGA-3’).
Extraction of the total RNA
The total RNA was isolated using the PureLink RNA mini kit (Ambion, Life Technologies, USA) according to the manufacturer’s instructions. Equal concentrations and volumes of the tissue sampled from different individuals were combined to produce four RNA pools, separated by organism and type of tissue. The samples were treated with RNAse-free DNAse (Invitrogen, CA, USA) to remove any DNA contaminants. The amount of RNA extracted was determined using a PicoDrop spectrophotometer (Picodrop, United Kingdom), and the quality, with a Bioanalyzer 2100 (Agilent Technologies). The integrity of the samples was confirmed by electrophoresis in 1.5% agarose gel. All the RNA samples were treated with RiboMinus (Invitrogen, CA, USA), according to the manufacturer’s instructions, for the selective depletion of the ribosomal RNA (rRNA) transcripts from the total RNA. The RNA samples were then stored at –80ºC prior to the pyrosequencing reaction.
Construction of the cDNA library
Four C. macropomum and tambacu libraries were established. Complementary DNA (cDNA) libraries were constricted in two steps, firstly, by the fragmentation of the total RNA and then the synthesis of the double-stranded cDNA. The initial quantity of RNA used in the fragmentation was approximately 600 ng/μ, with zinc chloride (ZnCl2) and Tris-HCL being added to initiate the fragmentation process. The integrity of the fragments was verified in the Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), using the RNA 6000 Pico kit. The double strands of the cDNA were then obtained using the cDNA Synthesis System kit (Roche), with the following steps: denaturation of the RNA by the addition of a randomic primer, synthesis of the first cDNA strand, synthesis of the second cDNA strand, and the purification of the double cDNA strands. The irregular ends of the cDNA fragments generated by this process were then repaired. The cDNA fragments were quantified using a TBS 380 QuantiFluor fluorometer (Promega, USA), according to the manufacturer’s instructions. The size of the cDNA fragments was verified in a bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) using the High Sensitivity DNA chip.
Amplification of the cDNA library and sequencing
The cDNA fragments of all the samples (C. macropomum and tambacu) were pyrosequenced in the 454 GS FLX Titanium platform (Roche, Branford, CT, USA). The emulsion PCR (emPCR) was based on the enrichment, purification, and preparation of the Pico Titer Plate (PTP), conducted according to the manufacturer’s instructions. All the libraries were sequenced five times, with each run using a PTP with two regions. The raw sequence reads have been submitted in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (Bioproject: PRJNA353388/ SRX2375124, SRX2375122, SRX2375120, SRX2375119).
Computational analysis
Pre-processing
The raw Sff files were converted into FastQ in Geneious v. 7.1.7 [26], and the adapters were removed. The ribosomal reads were then filtered using the default parameters and compared with the database available in SortMeRNA v. 2.0 [27]. The subsequent non-rRNA reads were separated by size (100 bps), quality (>20) and homopolymers (<12) in Mothur v. 1.35.1 [28].
De novo assembly
The contigs were assembled in Newbler v. 3.0 [29] and Mira v. 4.0.2 [30]. Both packages adopt the overlap-layout-consensus (OLC) algorithm [31]. The contigs obtained for each tissue and each assembler were clustered in CD-HIT-EST v. 4.6.4 [32] to remove redundant elements based on a minimum overlapping length of 60 bps and minimal overlapping similarity of 95%. A reference transcriptome was generated from the entire set of assembled contigs.
Assessment of transcriptome assembly completeness
The BUSCO v3 (Benchmarking Universal Single-Copy Orthologs) program [33] was used to evaluate transcriptome completeness. The transcriptomes were compared with a predefined set of eukaryote and metazoan single-copy orthologs from the OrthoDB v9 database [34], assuming a cut-off of 1E-3. This analysis was based on the complete database of the transcripts of all the samples (skin and muscle) from both organisms.
Functional annotation
The contigs were compared with non-redundant protein sequences of the Swissprot protein (Swiss-Prot) dataset using BlastX, assuming a cut-off of 1E-3 and similarity of at least 30%. The Swiss-Prot is a highly accurate protein database with high rates of annotation, reduced redundancy, and high levels of integration with other datasets [35]. The XML outputs from BlastX were annotated in Blast2GO v. 3.3 [36], being assigned to one of the three main categories of Gene Ontology (GO): Biological Processes (BP), Molecular Function (MF), and Cell Components (CC). The contigs were then mapped according to their known metabolic and molecular pathways in the vertebrates, using the Kyoto Encyclopedia of Genes and Genomes (KEGG) [37].
Differential expression
The total reads of each library were mapped in STAR 2.5 [38] based on the reference transcriptome (the entire set of assembled contigs). The individual counts of the transcripts identified in each tissue were obtained in HTSeq 0.6.1 [39], and were used as input to estimate the abundance of transcripts in RPKM (Reads per Kilobase per Million mapped reads) in the R statistical package. The read counts were normalized using the RPKM function of the EdgeR package [40]. The online software IDEG6 (p value <0.001) [41] was used to identify the differentially expressed genes (DEGs), confirmed by Fold-change log 2 (Log2FC) values ≤ -1, ≥ 1. The Heatmap graph was based on the RPKM values of the DEGs, as identified by the IDEG6 (≤ 0.001) and Log2FC values. The CIM function of the mixOmics package [42] was used to run the correlation analysis, and all transcripts that presented read numbers of zero in at least one library were removed from the analysis. The Volcano Plot was based on the significant p (≤ 0.001) and Log2FC (≤ -1, ≥ 1) values. The heatmaps were produced in Heatmap.2 [43] and the Volcano plots were obtained in Ggplot [44] from the R package.
Results and discussion
Genetic identification of the organisms
The morphological similarities between the C. macropomum and its hybrid forms have long been an impediment on the precise identification of the species reared in tropical aquaculture. Therefore, genetic markers are required to ensure the reliable identification of specimens. The markers used in the present study were the mitochondrial D-loop sequences and the multiplex PCR of the nuclear α-Tropomiosin gene. The multiplex PCR is widely used as a highly effective, low-cost tool for the rapid identification of interspecific hybrids and parent species [13, 45].
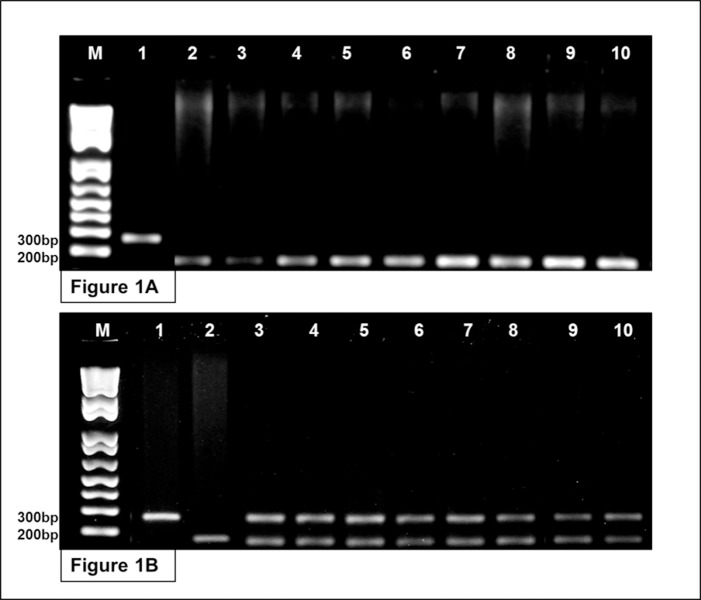
As the mitochondrial DNA is inherited maternally in most eukaryotes [46, 47], the sequencing of the D-loop confirmed that the female parents of all the specimens analyzed were C. macropomum (S1 Table). The hybrids were discriminated from the C. macropomum by multiplex PCR, using the parental species (C. macropomum and P. mesopotamicus) as control. The results of the PCR yielded distinct band profiles for P. mesopotamicus, with fragments of 300 base pairs (bps), and C. macropomum, which presented bands of only 200 bps. The multiplex PCR produced a single band of 200 bps in eight of the 16 specimens analyzed, confirming that they belong to C. macropomum (Fig 1A). The other eight specimens had two distinct bands (one of 300 bps and the other of 200 bps), confirming their identification as tambacu hybrids. The 300-bp fragment is inherited from the P. mesopotamicus father and the 200-bp fragment from the C. macropomum mother (Fig 1B).
Fig 1. Molecular identification of the organisms based on multiplex PCR of α–tropomiosin gene.
Fig 1A. Samples 1 and 2 represent the control species P. mesopotamicus (300 bps) and C. macropomum (200 bps), respectively. Samples 3–10 represent the pure C. macropomum individuals sampled in the present study. Fig 1B. Samples 1 and 2 represent the control species P. mesopotamicus (300 bps) and C. macropomum (200 bps), respectively. Samples 3–10 represent the hybrid tambacu individuals sampled in the present study.
Results of the sequencing and assembly
A total of 8,188,945 raw reads were obtained from the four libraries derived from the samples of the skin and muscle tissue of C. macropomum and the tambacu, using the Roche 454 platform (Table 1). After filtering, the remaining reads obtained from each type of tissue were assembled separately using MIRA and Newbler, providing a total of 572,760 contigs. The dataset was analyzed in CD-Hit-Est to remove redundant contigs and putative assembly errors, resulting in a final number of 400,845 contigs (Table 1). The use of two assemblers enriched both the number and the size of contigs, and is a successful, widely-used strategy [48, 49]. The contigs ranged in size from 200 to 49,961 bps. The largest contig was assembled from the C. macropomum muscle samples (S1 Fig). The N50 values for each type of tissue and other assembly metrics are shown in Table 1.
Table 1. De novo assembly and contigs obtained for each type of tissue in the C. macropomum and tambacu samples.
| Organism | C. macropomum (Skin) | tambacu (Skin) | C. macropomum (Muscle) | tambacu (Muscle) |
|---|---|---|---|---|
| Total Reads | 2,359,900 | 1,902,668 | 2,043,773 | 1,882,604 |
| Filtered Reads | 778909 | 430979 | 641023 | 843675 |
| Mean Quality | 32.4 | 32.82 | 30.74 | 31.08 |
| Average Length Reads | 442.5 | 363.54 | 409.82 | 401.16 |
| De novo assembly | NEWBLER MIRA | NEWBLER MIRA | NEWBLER MIRA | NEWBLER MIRA |
| Assembled Reads | 225,669 276,699 | 57,499 77,521 | 282,652 251,282 | 329,908 336,303 |
| Total Contigs | 167,404 94,624 | 62,468 26,030 | 42,235 31,030 | 92,284 56,685 |
| Largest Contig | 16,759 10,530 | 7,664 5,909 | 49,960 22,203 | 43,182 49,465 |
| N50 | 826 971 | 743 858 | 837 846 | 796 836 |
| Average Length | 817 910.5 | 753 776.8 | 833 768.5 | 817 786.8 |
| Contigs after CD-Hit-Est | 179,830 | 66,583 | 47,985 | 106,447 |
| Annotated Contigs | 18,200 | 7,932 | 11,316 | 20,875 |
Assessment of the completeness of the transcriptome
The completeness of the transcriptome assembly of the C. macropomum and tambacu samples was assessed through comparison with the Benchmarking Universal Single-Copy Orthologs (BUSCO) for the metazoan and eukaryote gene sets. The genes sets were classified according to BUSCO parameters (Table 2) as either Complete (C), Single (S), Duplicated (D), Fragmented (F) or Missing (M). In this analysis, including the total set of transcripts of C. macropomum and the tambacu, 41.1% of the complete genes were observed in comparison with the metazoan gene set, and 42.3% in comparison with the eukaryote set. If the fragmented genes are included here, the percentage of genes recovered increases to 75.5% in comparison with the metazoans and 77.9% for the eukaryotes. On the other hand, 24.55% of the genes were classified as missing in comparison with the metazoan set, and 22.1% in comparison with the eukaryote set, which may reflect biological innovations in the C. macropomum and tambacu [33]. Although this cannot be evaluated due to the lack of data on the complete genome of these organisms, or even this result may be a consequence of problems in the data assembled.
Table 2. BUSCO statistics for the completeness of the C. macropomum and tambacu transcriptome assembly in comparison with the metazoan and eukaryote gene sets.
| BUSCO | Metazoan Odb9 (%) | Eukaryote Odb9 (%) |
|---|---|---|
| Complete | 41.1 | 42.3 |
| Single | 28.9 | 29.4 |
| Duplicated | 12.2 | 12.9 |
| Fragmented | 34.4 | 35.6 |
| Missing | 24.5 | 22.1 |
Functional annotation
When the contigs from the present study were compared to the Swiss-Prot database using BlastX, 100,298 (25%) of the transcripts having significant hits. Of this total, 58,322 contigs were annotated (S2 Fig). Overall, 19.4% of the transcripts annotated in C. macropomum referred to the muscle, and 31.2% to the skin. In the tambacu 35.8% referred to the muscle and 13.8% to the skin. The top hits in BlastX revealed that C. macropomum and its hybrid shared transcripts with a number of other species of fish, amphibian, and mammal (S3 and S4 Figs).
Gene Ontology (GO)
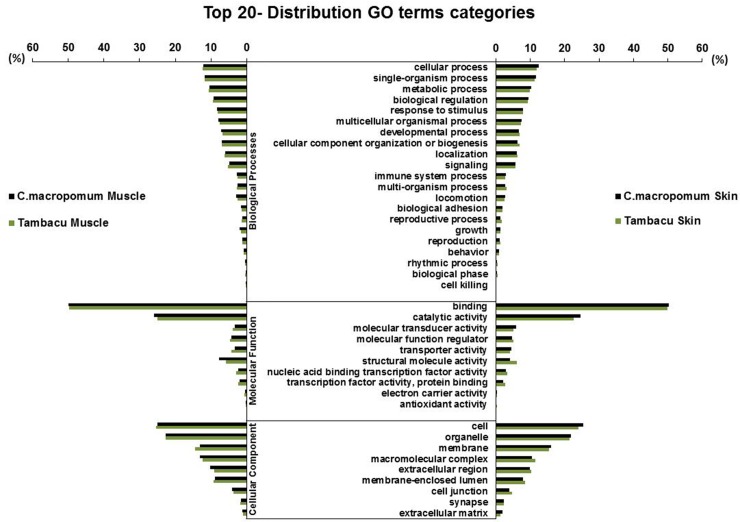
The Gene Oncology (GO) project provides a useful bioinformatics tool for the representation of genes or their products in different species and are especially informative for large datasets [50, 51]. In the present study, the GO analysis of the three principal domains, Biological Processes (BP), Molecular Function (MF), and Cell Components (CC), revealed a large number of functions for C. macropomum (15,634 GO terms) and tambacu (15,140 GO terms). The BP domain had the highest number of functions, with 10,491 (67.1% of the total) terms in the C. macropomum and 10,104 (66.7%) in the tambacu. This domain include a number of distinct systems, ranging from metabolic pathways to the physiological and behavioral features of an organism [52]. This domain was followed by MF, with 3,675 (23.5%) terms in the C.macropomum and 3,584 (23.7%) in the tambacu, while the CC had 1,469 (9.4%) in the C. macropomum and 1,452 (9.6%) in the tambacu (S5 Fig).
Similar results were obtained for the C. macropomum and tambacu in relation to the main functions observed in the BP, MF, and CC domains (Fig 2). The GO term “cellular process” (GO:0009987) was predominant in the BP domain (>12% in the four libraries), whereas in the MF domain, “binding” (GO:0005488) was the most dominant, with nearly half of the transcripts, followed by “catalytic activity” (GO:0003824), responsible for another quarter. In the case of the CC domain, the “cell” (GO:0005623) and “organelle” (GO:0043226) were the most common (45% of the transcripts in the four libraries). The distribution of the most expressed GO terms from each domain were similar to those reported in the transcriptomes of other vertebrate species [53–55].
Fig 2. Top 20 GO terms.
Processes with the highest number of transcripts within Biological Processes (BP), Molecular Function (MF) and Cell Components (CC) domains in the muscle and skin of C. macropomum and tambacu.
Kyoto Encyclopedia of Genes and Genomes (KEGG)
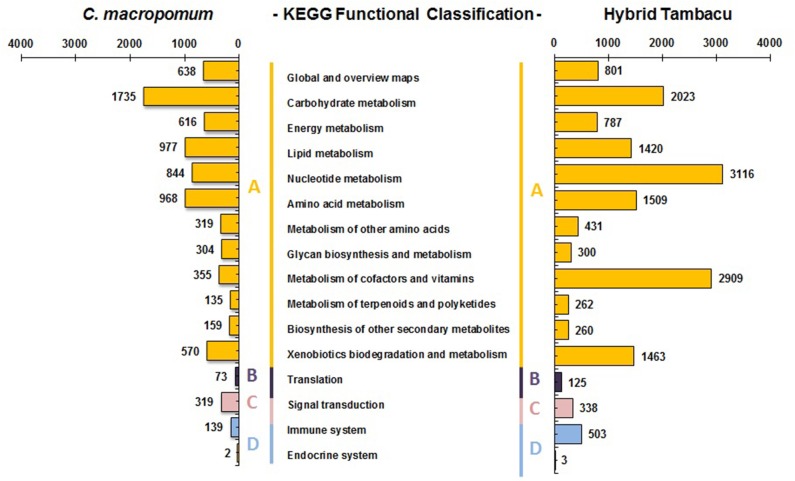
The KEGG analysis provides plots of the biochemical pathways, and is useful for the systematic analysis of gene function, focusing in particular on the gene and the molecular network [56]. In the present study, we combined the tissues from each organism, resulting in 132 signaling pathways for the C. macropomum and 133 for the tambacu, corresponding to 8276 (1337 enzymes) and 16,366 (1334 enzymes) sequences, respectively (S2 and S3 Tables). The pathways were classified in four categories: Metabolism, Genetic information processing, Environmental information processing, and Organismal system (Fig 3). The largest numbers of transcripts (7,620 in C. macropomum and 15,281 of those in the tambacu) were allocated to the Metabolism category. Within this category, a number of differences were found between the two organisms in the distribution of the subcategories. With 22.7% of the C. macropomum transcripts being involved with carbohydrate metabolism and 12.8% with lipid metabolism, whereas in the tambacu, 19.0% of the transcripts were involved with nucleotide metabolism and 17.7% with the metabolism of cofactors and vitamins.
Fig 3. Pathway assignment based on the Kyoto Encyclopedia of Genes and Genomes (KEGG).
(A) Metabolism categories, (B) Genetic information processing, (C) Environmental information processing, and (D) Organismal systems.
We identified some differences in the most common pathways in the C. macropomum and tambacu, although in both organisms, the purine metabolism pathways were predominant, with 689 transcripts in the C. macropomum, and 2850 in the tambacu (S6 and S7 Figs). Previous reports on the fish transcriptome produced similar results, such as those recorded for the hybrid pufferfish (Takifugu rubripes x Takifugu flavidus) [57], for example, and the closely-related species, Piaractus mesopotamicus [6] and Piaractus brachypomus [58].
Differential expression
The 454 FLX Genome sequencer (GS FLX) is suitable for the de novo sequencing of the transcriptome of non-model organisms [59–61], as confirmed in a number of previous studies [5, 6, 62]. As the organisms analyzed did not have a reference genome, we prepared a reference transcriptome, based on the entire set of assembled contigs. These sequences were mapped in the Star software, and counted in Htseq. Using rigorous parameters, we obtained a mapping index of 64.6% for C. macropomum, and 65.4% for the tambacu.
The present study provides the first comparative analysis of the transcriptomes of C. macropomum and the tambacu. Thus, based on the 400,845 contigs compiled in the four libraries, 21,986 genes were identified, including isophorms (S4 Table), with 16,632 being identified for C. macropomum and 14,835 for tambacu. The dataset from the muscle from both organisms comprised 13,951 genes, with 2,352 expressed only in the C. macropomum and 6,630 only in the tambacu. The dataset for the skin was composed of 16,857 genes, of which, 9,371 were expressed only in the C. macropomum and 2,772 only in the tambacu. The differences in the number of exclusive and shared genes support the hypothesis that the expressed transcripts depend on the development stage or genetic background of the sample specimens, given that genes may be expressed differentially in the same tissue [63, 64].
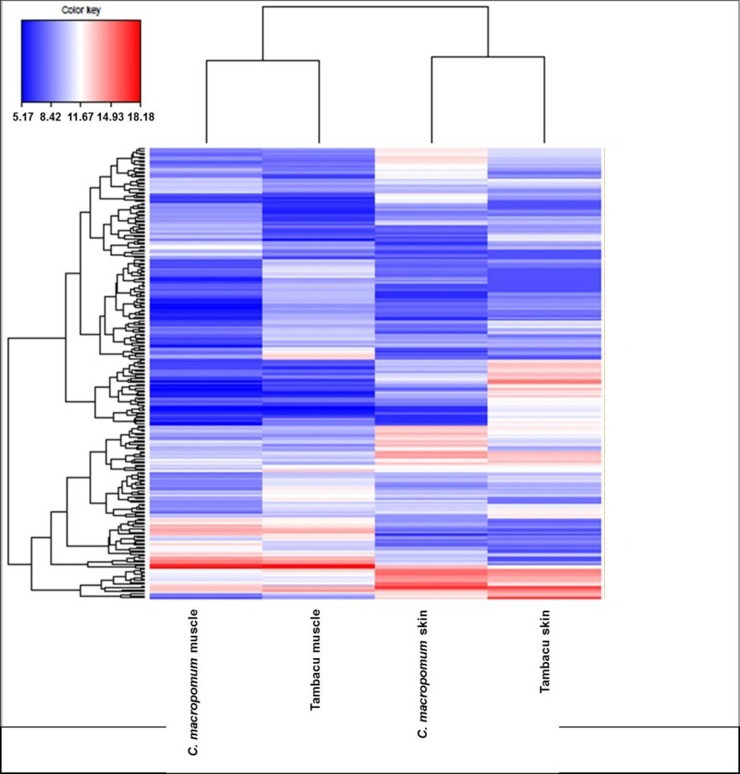
The results of the heatmap analysis (Fig 4) indicate the clustering of tissues, with one clade formed by the muscle transcriptome of the C. macropomum and tambacu, and the other by the skin samples of the two organisms. The similar size of branches in both clades indicates equivalent patterns of divergence in both tissues in each organism. Each library was also characterized by a number of different groups of distinct and overexpressed genes, considering only the DEGs with RPKM <100 (all the genes included in the analysis are shown in S5 Table). The IDEG6 analysis (p < 0,001) together with the Log2FC values (≥ 1 and ≤ -1) revealed a number of differentially-expressed genes (see the DEGs in S6 Table). A total of 168 up-regulated genes were found in the library of the C. macropomum muscle, 222 in the tambacu muscle, 95 in the C. macropomum skin and 115 in the tambacu skin (Figs 5 and 6). These results reinforce the differences found in gene expression among organs, tissues or even different types of cell from the same organ [65].
Fig 4. Heatmap graph.
Constructed with RPKM values of differentially expressed genes for C. macropomum and tambacu (P Value ≤ 0,001; Log2FC ≥ 1 e ≤ -1).
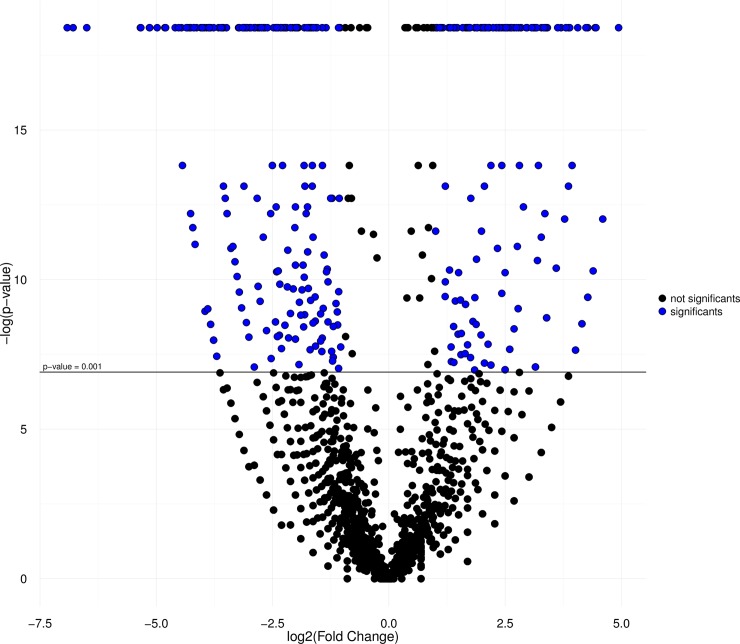
Fig 5. Volcano plot graph.
Comparison of C. macropomum muscle DEGs (P ≤0,001; Log2FC ≥1) and tambacu muscle (P ≤ 0,001; Log2FC: ≤ -1), in red color the transcripts with significant values and in color blue the no significant.
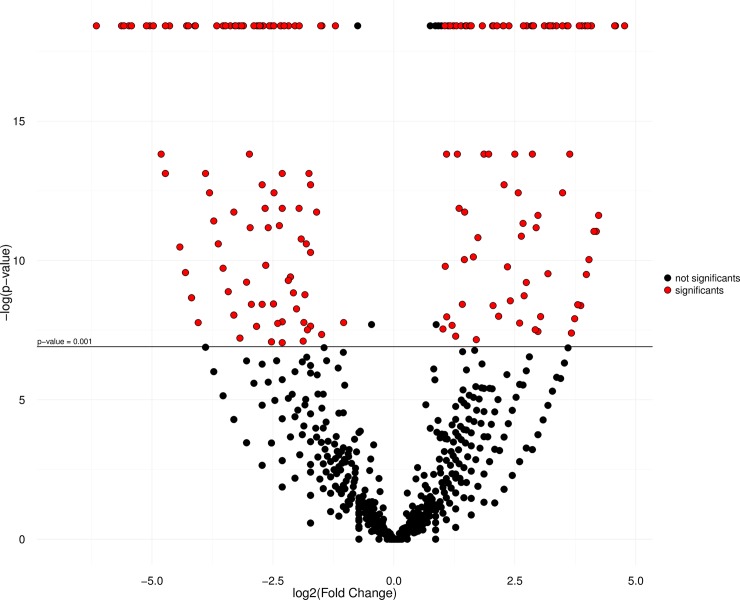
Fig 6. Volcano plot graph.
Comparison of C. macropomum skin DEGs (P ≤0,001; Log2FC ≥1) with tambacu skin (P ≤ 0,001; Log2FC ≤ -1), in red color the transcripts with significant values and in color black the no significant.
Genes expressed differentially in the muscle of the C. macropomum and tambacu
The largest contig with functional annotation (54,960 bps) was observed in the muscle of the C. macropomum, while the same transcript in the tambacu was slightly shorter, at 54,465 bps, but was nevertheless the most abundant protein (in reads) in this organism. This transcript refers to the gene involved in the synthesis of Titin (ttn), which is the largest and the most abundant protein in the skeletal and cardiac muscle of humans [66, 67]. In C. macropomum, the most expressed gene was the Myosin heavy chain fast skeletal muscle (myss), which is related to muscle contraction, and was also up-regulated in this species (Table 3).
Table 3. Some of the genes with the highest level of expression identified in the muscle and skin of C. macropomum and the tambacu.
| Genes | Length | Over-Express | tissue | RPKM C.macropomum | RPKM tambacu | FC | Log2FC | P Value |
|---|---|---|---|---|---|---|---|---|
| Myosin heavy chain, fast skeletal muscle | 7110 | C. macropomum | Muscle | 12424,47 | 5090,61 | 2,44 | 1,28 | <0,001 |
| Parvalbumin beta | 2857 | C. macropomum | Muscle | 10822,37 | 832,01 | 13,007 | 3,71 | <0,001 |
| Beta-enolase | 1650 | C. macropomum | Muscle | 3422,18 | 722,15 | 4,738 | 2,24 | <0,001 |
| Actin, alpha skeletal muscle | 1135 | C. macropomum | Muscle | 1889,28 | 322,40 | 5,859 | 2,55 | <0,001 |
| Myosin-2 | 1379 | C. macropomum | Muscle | 768,64 | 35,08 | 21,905 | 4,45 | <0,001 |
| ADP/ATP translocase 1 | 1398 | tambacu | Muscle | 146,74 | 1213,59 | 0,12 | -3,04 | <0,001 |
| Elongation factor 1-alpha | 2219 | tambacu | Muscle | 356,60 | 962,20 | 0,37 | -1,43 | <0,001 |
| 4F2 cell-surface antigen heavy chain | 2078 | tambacu | Muscle | 21,15 | 852,84 | 0,02 | -5,33 | <0,001 |
| Alpha-crystallin B chain | 2344 | tambacu | Muscle | 20,83 | 845,08 | 0,02 | -5,34 | <0,001 |
| Tripartite motif-containing protein 54 | 1689 | tambacu | Muscle | 52,84 | 668,71 | 0,07 | -3,66 | <0,001 |
| LINE-1 retrotransposable element ORF2 protein | 3900 | C.macropomum | Skin | 558,90 | 256,92 | 2,17 | 1,12 | <0,001 |
| Collagen alpha-1 (I) chain | 5865 | C.macropomum | Skin | 511,16 | 33,03 | 15,4 | 3,95 | <0,001 |
| Collagen alpha-3 (VI) chain | 9460 | C.macropomum | Skin | 334,63 | 28,08 | 11,9 | 3,57 | <0,001 |
| LINE-1 reverse transcriptase homolog | 3533 | C.macropomum | Skin | 307,53 | 117,51 | 2,61 | 1,38 | <0,001 |
| Cation channel sperm-associated protein | 4478 | C.macropomum | Skin | 85,37 | 9,88 | 8,63 | 3,10 | <0,001 |
| Keratin, type II cytoskeletal 8 | 2829 | tambacu | Skin | 160,02 | 1473,49 | 0,11 | -3,21 | <0,001 |
| Keratin, type I cytoskeletal 13 | 1687 | tambacu | Skin | 47,70 | 826,93 | 0,05 | -4,11 | <0,001 |
| Ictacalcin | 809 | tambacu | Skin | 111,91 | 650,07 | 0,17 | -2,53 | <0,001 |
| Desmoplakin | 2927 | tambacu | Skin | 18,33 | 455,80 | 0,04 | -4,63 | <0,001 |
| Keratin, type I cytoskeletal 50 kDa | 1992 | tambacu | Skin | 10,10 | 333,48 | 0,03 | -5,04 | <0,001 |
Of the 168 up-regulated genes in the muscle of C. macropomum, those of the myosin gene family are the most prominent, including Myosin-1 (myh1), Myosin-2 (myh2), Myosin-3 (myh3), Myosin-4 (myh4), Myosin-7 (myh7), Myosin-7B (my7b), Myosin-8 (myh8), and Myosin-13, myh13 (Table 3, S6 Table). This gene family encodes motor proteins, which are responsible for the contraction mechanism, as well as being involved in several mechanochemical processes in eukaryotes [68, 69]. In addition to this gene family, Actin (acts/acta1) was also expressed prominently in the muscle, and is particularly active in the eukaryote cell, where it has a synergistic effect with myosin in the control of mobility and muscle contraction [70], as well as the genes that encode Myomesin 1 (myom1) and Myomesin 2 (myom2), the main components of M-line [71] (Table 3, S6 Table). It is interesting to note that all these up-regulated genes in C. macropomum are related to muscular contraction. This may be related to the fact that this species is rheophilic, and needs to make an enormous effort to swim upstream, against the current, to mature sexually. The individuals raised in captivity (as those used in the present study) can to mature sexually, although the final stage of the spawning process does not occur, given the absence of the external factors necessary to trigger the reproductive process [7].
The genes responsible for the expression of catalytic proteins were also abundant in the DEGs of the muscle of the C. macropomum, including the enzymes Beta Enolase (enob/ eno3), Triosephosphate Isomerase B (tpisb/tip1b), NAD(P)H dehydrogenase (quinone) 1 (nqo1), Enolase (eno), and Phosphoglucomutase-1 (pgm1), most of which are related to the synthesis or degradation of glucose and the metabolism of carbohydrates [72–74] (Table 3, S6 Table). This finding may account for the predominance of the carbohydrate metabolism subcategory identified in the KEGG analysis for C. macropomum.
The results obtained for the transcriptome of the tambacu muscle included several up-regulated genes related to processes such as aminoacid metabolism, stress, and catalysis. In the KEGG analysis of the tambacu muscle, nucleotide metabolism was the most common subcategory (involving 19% of the transcripts) in the “metabolism” category. This finding can be accounted for by the presence of a number of differentially-expressed genes related to the metabolism of aminoacids, such as Elongation factor 1-alpha (eef1a), 4F2 cell-surface antigen heavy chain (4f2/ slc3a2), and Large neutral amino acids transporter small subunit 4 (lat4/ slc43a2) [75–77] (Table 3, S6 Table). The DEGs observed in the muscle of the tambacu also included genes of the ubiquitin family and similar genes, such as Ubiquitin-40S ribosomal protein S27a (rps27a), F-box only protein 32 (fbxo32), AN1-type zinc finger protein 5 (zfand5) (Table 3, S6 Table). These genes are involved primarily in the regulation of proteins [78].
Genes of the Heat shock protein (HSP) family were also prominent in the up-regulated genes of the muscle of the tambacu: Heat shock protein beta-1 (hspb1), Heat shock protein beta-11 (hspb11/hspbb), Heat shock 70 kDa protein (hsp70), Heat shock 70 kDa protein 1 (hsp71), Heat shock protein beta-6 (hspb6), Heat shock protein beta-8 (hspb8) (S6 Table). The expression of this protein family is related mainly to environmental and physiopathological stress [79, 80]. The Alpha-crystallin B (cryab) proteins are also prominent in this group (Table 3), being characterized by their presence in a number of different types of tissue, including the skeletal muscle [81, 82]. Like other HSPs, these proteins act as molecular chaperones to prevent the incorrect association and aggregation of the polypeptide chain [83, 84]. The presence of a number of HSPs with prominent over-expression in the muscle of tambacu is noteworthy, given that the expression of these genes is usually a response to stress factors, including environmental conditions and disease [79, 80, 85]. The marked expression of the genes related to these proteins potentially indicates environmental effects and adaptive processes in the hybrid tambacu. In fact, the HSPs are helpful for the repair of protein damages, given that over-expression is usually sufficient to protect the cell from exposure to stressful conditions, such as high temperatures [80, 86].
Some transcripts related to processes such as catalytic action, defense, and oxidative stress were also relatively common in the DEGs of the tambacu muscle, including AMP desaminase 3 (ampd3), AMP desaminase 1 (ampd1) Phosphoglycerate mutase 2 (pgam2), Sestrin-1 (Sesn-1), and Thioredoxin-interacting protein (txnip) [87–89] (S6 Table).
Genes expressed differentially in the skin of the C. macropomum and tambacu
The skin is a complex type of tissue that plays a key role in sensorial activities, thermoregulation, hormone metabolism, and defense [20, 90, 91]. In addition to these functions, the morphological similarities between the C. macropomum and the tambacu also justifies the analysis of this tissue. Overall, 210 DEGs were identified in the skin of C. macropomum and tambacu.
In the skin of the C. macropomum, the transcripts with the largest number of mapped reads corresponded to the transposable elements: Transposable element Tcb1 transposase (tcb1), Transposable element Tc1 transposase (tc1a), and Transposable element Tcb2 transposase (tcb2). These elements are DNA segments capable of moving through the genome of a cell (“jumping genes”), thus affecting the evolution and genome composition of plants and animals, inducing processes ranging from pathologies to genetic differences in these organisms [92–94]. As in the muscle, the genes that encode collagen proteins were also found to be over-expressed in the skin of C. macropomum, including: Collagen alpha-1(I) chain (co1a1), Collagen alpha-3(VI) chain (co6a3) and Collagen alpha-2(I) chain (co1a2). These proteins are abundant in animals and are important for the molecular architecture and the shape and mechanical properties of tissues [95, 96].
Another important gene family, that should be mentioned, is the Cation channel sperm-associated protein (catsper1/ctsr1 and catsper3/ctsr3). Despite presenting RPKM values lower than 100 (Table 3, S6 Table), this gene group was up-regulated in the skin of C. macropomum. However, the contigs equivalent to these transcripts were not assembled in the tambacu. The proteins encoded by this gene form a complex ion channel that permits the Ca2+ to enter the flagellum of the spermatozoa, which ensures their hyperactivation and plays a key role in male fertility [97]. The lack of this transcript in the tambacu may be related to the infertility or subfertility of these hybrids. Although some authors have observed that tambacu males produce small amounts of spermatozoa following abdominal pressure, which indicates that these fish can be crossed with their parent species (unpublished data), it remains unclear whether the tambacu is effectively fertile. In addition, the presence of up-regulated transcripts of catsper1 and catsper3 in the C. macropomum skin may represent a potential genetic marker for sex differentiation of the specie, which would be important, given the lack of sexual dimorphism in these fish during non-breeding periods, as occurred in the present study.
With regard to the differential expression of genes in the skin of the tambacu, the transcript with the largest number of mapped reads was related to the keratin, type II cytoskeletal 8 (krt8) protein, which was also present in the DEGs. In fact, other transcripts that encode proteins of this large keratin family (classes I and II) were also found in the DEGs of the tambacu skin (Table 3, S6 Table). The products of this gene family are structural proteins of the epithelial cells, which are expressed differentially according to the cell layer and tissue. The functions of these proteins have not yet been determined fully, but they are known to be involved in a number of processes, including structural support, cytoarchitecture, the response to stress, the regulation of signaling pathways related to apoptosis and protein synthesis, the distribution of organelles, and tissue repair and proliferation [98–100].
In contrast with the pattern observed in the C. macropomum, only a single gene of the collagen family–the collagen alpha-1(XVII) chain (Col17a1)–was observed in the DEGs of the tambacu skin. This transmembrane protein is a structural component of the hemidesmosomes in the epithelial cells and is associated with the keratinocytes [101]. One other important gene is the dsp, which encodes desmoplakin (Table 3). This is a key desmosome protein, which plays a major role in the attachment, assembly and stabilization of intermediate filaments, in addition to interacting with keratin proteins [102, 103].
Some authors have suggested that hybrids are more resistant and grow faster than their parent species [11, 104]. However, few studies have demonstrated any actual advantages in terms of increased productivity, and in fact, some data indicate that hybrids may be more susceptible to parasites [105, 106]. In the present study, major differences were found in the specificity and levels of gene expression in the muscle and skin of the C. macropomum and tambacu, which may contribute to the identification of the specific genes involved in the immunity, growth and fertility of these organisms. These findings raise a number of important questions that must be settled in order to confirm whether hybrids have any real advantages over their parent species, and whether they constitute a threat to the stocks of native species. We would further emphasize here the various up-regulated genes linked to stress found in the tambacu, which may be relevant to the farming of this hybrid, given that the stress of the captive environment may have a negative effect on the development of the fish. Given this, it will be very important to identify the factors that provoke this condition, not only for the organism itself, but also fish farming (the region in which this hybrid is farmed should also be evaluated).
Conclusion
This is the first comparative analysis of the C. macropomum and its hybrid form, the tambacu. The results indicated that the NGS platform used in the study is a powerful tool for the identification of genes and molecular markers in non-model species, in addition to confirming that the combination of distinct assembly methods can enrich the dataset considerably. Despite of the lack of a reference genome, we obtained a satisfactory number of transcripts with functional annotation, associated typically with biological processes, with the skin presenting the greatest number of GO terms. The KEGG analysis also identified similarities in the metabolic pathways of the two organisms, with several transcripts in the metabolism category. There was a degree divergence in the number of exclusive genes, and the levels of gene expression and function between the C. macropomum and tambacu tissues. However, a significant number of genes remained uncharacterized, and are also of particular interest for future studies on the physiology, conservation, and genetic improvement of the native species, as well as the management of hybrid stocks. This research is thus an important contribution to the investigation of genes that contribute to potential advantages or disadvantages in the productivity of the hybrids, their implications for the conservation and management of C. macropomum, and provide a valuable tool for future research in functional genomics.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
Comparative distribution of the GO terms and the numbers of transcripts involved in Biological Processes (BP), Molecular Function (MF), and Cell Components (CC) in the C. macropomum and tambacu.
(TIF)
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(CSV)
(XLSX)
Acknowledgments
We are grateful to the Center for Technological Innovation of the Evandro Chagas Institute for the formal partnership that supported this study. Bionorte Project and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the PhD scholarship granted. The Cristiana Maciel for the availability of her laboratory to perform RNA extraction.
Data Availability
All relevant data are within the paper and its Supporting Information files. Raw reads data that support the findings of this study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (Bioproject: PRJNA353388/ Acession number: SRX2375124, SRX2375122, SRX2375120, and SRX2375119).
Funding Statement
The study was funded by the Center of Technological Innovation (CIT), Evandro Chagas Institute (IEC) and National Council of Scientific and Technological Development (CNPq) (Bionorte Project: 550939/2010-5). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. 10.1016/j.ygeno.2008.07.001 [DOI] [PubMed] [Google Scholar]
- 2.Morozova O, Hirst M, Marra MA. Applications of New Sequencing Technologies for Transcriptome Analysis. Annu Rev Genomics Hum Genet. 2009;10:135–151. 10.1146/annurev-genom-082908-145957 [DOI] [PubMed] [Google Scholar]
- 3.Datta S, Nettleton D. Stattisticall Analysis of Next Generation Sequencing Data. Springer: New York; 2014. [Google Scholar]
- 4.Shen XY, Kwan HY, Thevasagayam NM, Prakki SRS, Kuznetsova IS, Ngoh SY, et al. The first transcriptome and genetic linkage map for Asian arowana. Mol Ecol Resour. 2014;14:622–635. 10.1111/1755-0998.12212 [DOI] [PubMed] [Google Scholar]
- 5.Thevasagayam NM, Sridatta PSR, Jiang J, Tong A, Saju JM, Kathiresan P, et al. Transcriptome Survey of a Marine Food Fish: Asian Seabass (Lates calcarifer). Mar Sci Eng. 2015;3:382–400. [Google Scholar]
- 6.Mastrochirico-Filho VA, Hata ME, Sato LS, Jorge PH, Forest F, Rodriguez MV, et al. SNP discovery from liver transcriptome in the fish Piaractus mesopotamicus. Conserv Genet Resour. 2016;8:109–114. [Google Scholar]
- 7.Lima CA, Goulding M. Os Frutos do Tambaqui: Ecologia, Conservação e Cultivo na Amazônia. Tefé, AM: Sociedade Civil de Mamiraua; 1998. [Google Scholar]
- 8.Ministério da Pesca e Aquicultura. Boletim Estatístico da Pesca e Aquicultura 2011. Brasil: Brasília; 2014. [Google Scholar]
- 9.Pinheiro MHP, Silva JWB, Nobre MIS, Pinheiro FA. Cultivo do Híbrido Tambaqui, Colossoma macropomum CUVIER, 1818, com a Pirapitinga C. brachypomum Cuvier, 1818, na Densidade de 5.000 Peixes/ha. Ciênc Agron. 1991;22:77–87. [Google Scholar]
- 10.Giuseppe M. Retrocruce de hembras híbridos (F1) (Colossoma macopomum X Piaractus mesopotamicus) com machos de lãs especies parentales. Mem Soc Ci Nat La Salle. 1994;54:9–13. [Google Scholar]
- 11.Melo JSC, Pereira JA. Crescimento do híbrido Tambacu (Fêmea de Colossoma macropomum X Macho de Piaractus mesopotamicus) em criação intensiva. Boletim técnico CEPTA. 1994;7:59–75. [Google Scholar]
- 12.Calcagnotto D, Almeida-Toledo LF, Bernardino G, Toledo-Filho SA. Biochemical genetic characterization of F1 reciprocal hybrids between neotropical pacu (Piaractus mesopotamicus) and tambaqui (Colossoma macropomum) reared in Brazil. Aquaculture. 1999;174:51–57. [Google Scholar]
- 13.Gomes F, Schneider H, Barros C, Sampaio D, Hashimoto D, Porto-Foresti F, et al. Innovative molecular approach to the identification of Colossoma macropomum and its hybrids. An Acad Bras Cienc. 2012;84:517–525. [DOI] [PubMed] [Google Scholar]
- 14.Prado-Lima M, Val AL. Transcriptomic Characterization of Tambaqui (Colossoma macropomum, Cuvier, 1818) Exposed to Three Climate Change Scenarios. PLoS ONE. 2016;11:e0152366 10.1371/journal.pone.0152366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes F, watanabe L, Nozawa S, Oliveira L, Cardoso J, Vianez J, et al. Identification and characterization of the expression profile of the microRNAs in the Amazon species Colossoma macropomum by next generation sequencing. Genomics. 2017;67–74. [DOI] [PubMed] [Google Scholar]
- 16.Martínez JG, Machado VN, Caballero-Gaitán SJ, Santos MF, Alencar RM, Escobar MD, et al. SNPs markers for the heavily overfished tambaqui Colossoma macropomum, a Neotropical fish, using next-generation sequencing-based de novo genotyping. Conserv Genet Resour. 2017;9:29–33. [Google Scholar]
- 17.Nunes JS, Liu S, Pértille F, Perazza CA, Villela OS, De Almeida-Val VF, et al. Large-scale SNP discovery and construction of a high-density genetic map of Colossoma macropomum through genotypingby- sequencing. Sci Rep. 2017;7:46112 10.1038/srep46112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown TA. Genomes 2nd edition Oxford: Department of Biomolecular Science; 2002. [Google Scholar]
- 19.Goetz FW, MacKenzie S. Functional genomics with microarrays in fish biology and fisheries. Fish and Fish. 2008;9:378–395. [Google Scholar]
- 20.Elliott DG. Integumentary system In: Ostrander GK, editors. The laboratory fish, 1st edn New York: Academic Press; 2000. p. 271–306. [Google Scholar]
- 21.Micallef G, Bickerdike R, Reiff C, Fernandes JMO, Bowman AS, Martin SAM. Exploring the Transcriptome of Atlantic Salmon (Salmo salar) Skin, a Major Defense Organ. Mar Biotechnol. 2012;14:559–569. 10.1007/s10126-012-9447-2 [DOI] [PubMed] [Google Scholar]
- 22.Palstra AP, Beltran S, Burgerhout E, Brittijn AS, Magnoni LJ, Henkel CV, et al. Deep RNA Sequencing of the Skeletal Muscle Transcriptome in Swimming Fish. PLoS ONE. 2013;8:e53171 10.1371/journal.pone.0053171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murgas LD, Drumond MM, Pereira GM, Felizardo VO. Cycle manipulation and reproductive efficiency in native species of freshwater fish. Rev Bras Reprod Anim Supl. 2009;6:70–76. [Google Scholar]
- 24.Sambrook J, Russel DW. Molecular cloning: a laboratory manual. New York: Cold spring harbor laboratory press; 2001. [Google Scholar]
- 25.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 26.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopylova E, Noé L, Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;1:1–7. [DOI] [PubMed] [Google Scholar]
- 28.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platformindependent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Müller WE, Wetter T, et al. Using the miraEST Assembler for Reliable and Automated mRNA Transcript Assembly and SNP Detection in Sequenced ESTs. Genome Res. 2004;14:1147–1159. 10.1101/gr.1917404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–27. 10.1016/j.ygeno.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- 34.Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, et al. OrthoDB v9.1: cataloguing evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. OUP accepted manuscript. Nucleic Acids Res. 2016;45:1–15. 10.1093/nar/gkw1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bairoch A, Apweiller R. The Swiss-Prot protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gotz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, et al. High-throughput functional annotation and data mining with the blast2GO suíte. Nucleic Acids Res. 2008;36:3420–3435. 10.1093/nar/gkn176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriya Y, Itoh M, Okuda S. KAAS: Na automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq- a Python framework to work with high-throughput sequencing data. Bioinformatics. 2014;1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12(2):159–62. 10.1152/physiolgenomics.00096.2002 [DOI] [PubMed] [Google Scholar]
- 42.González I, Cao KL, Davis MD, Déjean S. Insightful graphical outputs to explore relationships between two ‘omics’ data sets. BioData Min. 2013;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WA, Lumley T, et al. gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1. 2016.
- 44.Wickham H. ggplot2: Elegant Graphics for Data Analysis Springer-Verlag; New York, 2009. [Google Scholar]
- 45.Hashimoto DT, Mendonça FF, Senhorini JA, Jehud B, Oliveira C, Foresti F, et al. Identification of hybrids between Neotropical fish Leporinus macrocephalus and Leporinus elongates by PCR-RFLP and multiplex-PCR: Tools for genetic monitoring in aquaculture. Aquaculture. 2010;298:346–349. [Google Scholar]
- 46.Moritz C, Dowling TE, Brown WM. Evolution of animal mitochondrial DNA: relevance for population biology and systematic. Ann Rev Ecol Syst. 1987;18:269–292. [Google Scholar]
- 47.Parker PG, Snow AA, Schug MD, Booton GC, Fuerst PA. What molecules can tell us about populations: choosing and using a molecular marker. Ecology. 1998;79:361–382. [Google Scholar]
- 48.Kumar S, Blaxter ML. Comparing de novo assemblers for 454 transcriptome data. BMC Genomics. 2010;11:571 10.1186/1471-2164-11-571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer B, Martini P, Biscontin A, Pitt C, Romualdi C, Teschke M, et al. Pyrosequencing and de novo assembly of Antarctic krill (Euphausia superba) transcriptome to study the adaptability of krill to climate-induced environmental changes. Mol Ecol Resour. 2015;15:1460–1471. 10.1111/1755-0998.12408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. 10.1093/bioinformatics/bti610 [DOI] [PubMed] [Google Scholar]
- 52.Thomas PD, Mi H, Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Cell Biol. 2007;11:4–11. [DOI] [PubMed] [Google Scholar]
- 53.Salem M, Rexroad CE, Wang J, Thorgaard, Yao J. Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics. 2010;11:564 10.1186/1471-2164-11-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y, Zhang S, Xu J, Feng J, Mahboob S, Al-Ghanim KA, et al. Comparative Transcriptome Analysis Reveals the Genetic Basis of Skin Color Variation in Common Carp. PLoS ONE. 2014;9:e108200 10.1371/journal.pone.0108200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng T, Pang C, Lu X, Zhu P, Duan A, Tan Z, et al. De Novo Transcriptome Assembly of the Chinese Swamp Buffalo by RNA Sequencing and SSR Marker Discovery. PLoS ONE. 2016;11(1):e0147132 10.1371/journal.pone.0147132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao Y, Zhang H, Gao Q, Wang L, Zhang F, Siva VS, et al. Transcriptome Analysis of Artificial Hybrid Pufferfish Jiyan-1 and Its Parental Species: Implications for Pufferfish Heterosis. Plos One. 2013;8:e58453 10.1371/journal.pone.0058453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jorge PH, Mastrochirico-Filho VA, Hata MA, Mendes NJ, Ariede RB, Freitas MV, et al. Genetic Characterization of the Fish Piaractus brachypomus by Microsatellites Derived from Transcriptome Sequencing. Front Genet. 2018;9:46 10.3389/fgene.2018.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vera C, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol. 2008;17:1636–1647. 10.1111/j.1365-294X.2008.03666.x [DOI] [PubMed] [Google Scholar]
- 60.Meyer E, Aglyamova GA, Wang S, Buchanan-Carter J, Abrego D, Colbourne KJ, et al. Sequencing and de novo analysis of a coral larval transcriptome using 454 GSFlx. BMC Genomics. 2009;10:219 10.1186/1471-2164-10-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Neil ST, Dzurisin JDK, Carmichael RD, Lobo NF, Emrich SJ, Hellmann JJ. Population-level transcriptome sequencing of nonmodel organisms Erynnis propertius and Papilio zelicaon. BMC Genomics. 2010;11:310 10.1186/1471-2164-11-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu Y, Huang M, Wang W, Guan J, Kong J. Characterization of gonadal transcriptomes from the turbot (Scophthalmus maximus). Genome. 2016;59:1–10. 10.1139/gen-2014-0190 [DOI] [PubMed] [Google Scholar]
- 63.Adams J. Transcriptome: Connecting the genome to gene function. Nature Education. 2008;1:195. [Google Scholar]
- 64.Cao D, Kocabas A, Ju Z, Karsi A, Li P, Patterson A, et al. Transcriptome of channel catfish (Ictalurus punctatus): initial analysis of genes and expression profiles of the head kidney. Anim Genet. 2011;32:169–188. [DOI] [PubMed] [Google Scholar]
- 65.Alvarez M, Schrey AW, Richards CL. Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Mol Ecol. 2015;24:710–725. 10.1111/mec.13055 [DOI] [PubMed] [Google Scholar]
- 66.Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, et al. The Complete Gene Sequence of Titin, Expression of an Unusual -700-kDa Titin Isoform, and Its Interaction with obscurin Identify a Novel Z-Line to I-Band Linking System. Circ Res. 2001;89:1065–1072. [DOI] [PubMed] [Google Scholar]
- 67.Sanger JW, Sanger JM. Fishing out proteins that bind to titin. J Cell Biol. 2001;154:21–24. 10.1083/jcb.200106072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hirayama Y, Watabe S. Structural differences in the crossbridge head of temperature associated myosin subfragment-1 isoforms from carp fast skeletal muscle. Eur J Biochem. 1997;246:380–387. [DOI] [PubMed] [Google Scholar]
- 69.Berg J, Powell BC, Cheney RE. A Millennial Myosin Census. Mol Biol Cell. 2001;12:780–794. 10.1091/mbc.12.4.780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sparrow JC, Nowak KJ, Hayley JD, Beggs AH, Walgren-Pettersson C, Romero N, et al. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul Disord. 2003;13:519–531. [DOI] [PubMed] [Google Scholar]
- 71.Porter JD, Merriam AP, Gong B, Kasturi S, Zhou X, Hauser KF, et al. Postnatal suppression of myomesin, muscle creatine kinase and the M-line in rat extraocular muscle. J Exp Biol. 2003;206:3101–3112. [DOI] [PubMed] [Google Scholar]
- 72.Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D. Structure of the Triosephosphate Isomerase Phosphoglycolohydroxamate Complex: Na Analogue of the Intermediate on the Reaction Pathway. Biochemistry. 1991;30:5821–5826. [DOI] [PubMed] [Google Scholar]
- 73.Pancholi V. Multifunctional a-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.StojKovic T, Vissing J, Petit F, Piraud M, Orngreen MC, Andersen G, et al. Muscle Glycogenosis Due to Phosphoglucomutase 1 Deficiency. N Engl J Med. 2009;361:425–427. 10.1056/NEJMc0901158 [DOI] [PubMed] [Google Scholar]
- 75.Condeelis J. Elongation factor 1α, translation and the cytoskeleton. Trends Biochem Sci. 1995;20:169–170. [DOI] [PubMed] [Google Scholar]
- 76.Kanai Y, Segawa H, Miyamoto KI, Uchino H, Takeda E, Endou H. Expression Cloning and Characterization of a Transporter for Large Neutral Amino Acids Activated by the Heavy Chain of 4F2 Antigen (CD98). J Biol Chem. 1998;273:23629–23632. [DOI] [PubMed] [Google Scholar]
- 77.Guetg A, Mariotta L, Bock L, Herzog B, Fingerhut R, Camargo SMR, et al. Essential amino acid transporter Lat4 (Slc43a2) is required for mouse development. J Physiol. 2015;593:1273–1289. 10.1113/jphysiol.2014.283960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. 10.1016/j.bbamcr.2004.09.019 [DOI] [PubMed] [Google Scholar]
- 79.Morimoto RI. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. [DOI] [PubMed] [Google Scholar]
- 80.Morimoto RI. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. [DOI] [PubMed] [Google Scholar]
- 81.Dubin RA, Wawrousek EF, Piatigorsky J. Expression of the Murine αB-Crystallin Gene Is Not Restricted to the Lens. Mol Cell Biol. 1989;9:1083–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jeanpierre C, Austruy E, Delattre O, Jones C, Junien C. Subregional physical mapping of an αB-crystallin sequence and of a new expressed sequence DllS877E to human llq. Mamm Genome. 1993;4:104–108. [DOI] [PubMed] [Google Scholar]
- 83.Jong WW, Leunissen JAM, Voorter CEM. Evolution of the a-Crystallin / Small Heat-Shock Protein Family. Mol Biol Evol. 1993;10:103–126. 10.1093/oxfordjournals.molbev.a039992 [DOI] [PubMed] [Google Scholar]
- 84.Derham BK, Harding JJ. α-Crystallin as a Molecular Chaperone. Prog Retin Eye Res. 1999;18:463–509. [DOI] [PubMed] [Google Scholar]
- 85.Wu C. Heat Shock transcription factors: structure and regulation. Annu Rev Cell Dev. 1995;11:441–469. [DOI] [PubMed] [Google Scholar]
- 86.Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactiv ation of damaged proteins. Annu Rev Genet. 1993;27:437–496. 10.1146/annurev.ge.27.120193.002253 [DOI] [PubMed] [Google Scholar]
- 87.Buckbinder L, Talbott R, Seizinger BR, Kley N. "Gene regulation by temperature-sensitive p53 mutants: identification of p53 response genes". Proc Nat Acad Sci U S A. 1994;91:10640–10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hancock CR, Brault JJ, Terjung RL. Protecting the cellular energy State during contractions: Role of Amp Deaminase. J Physiol Pharmacol. 2006;57:17–29. [PubMed] [Google Scholar]
- 89.Zhou R, Tardivel A, Thorens B, Choi I, Tschopp J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat Immunol. 2010;11:136–141. 10.1038/ni.1831 [DOI] [PubMed] [Google Scholar]
- 90.Guellec DL, Morvan-Dubois G, Sire JY. Skin development in bony fish with particular emphasis on collagen deposition in the dermis of the zebrafish (Danio rerio). Int J Dev Biol. 2004;48:217–231. 10.1387/ijdb.031768dg [DOI] [PubMed] [Google Scholar]
- 91.Rakers S, Gebert M, Uppalapati S, Meyer W, Maderson P, Sell AF, et al. ‘Fish matters’: the relevance of fish skin biology to investigative dermatology. Exp Dermatol. 2010;19:313–324. 10.1111/j.1600-0625.2009.01059.x [DOI] [PubMed] [Google Scholar]
- 92.Prak ET, Kazazian HH. Mobile elements and the human genome. Nat Rev Genet. 2000;1:134–144. 10.1038/35038572 [DOI] [PubMed] [Google Scholar]
- 93.Biémont C, Vieira C. Junk DNA as an evolutionary force. Nature. 2006;443:521–524. 10.1038/443521a [DOI] [PubMed] [Google Scholar]
- 94.Venner S, Feschotte C, Biémont C. Transposable elements dynamics: toward a community ecology of the genome. Trends Genet. 2009;25:317–323. 10.1016/j.tig.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kadler K. Extracellular matrix 1: Fibril-forming collagens. Protein Profile. 1995;2:491–619. [PubMed] [Google Scholar]
- 96.Ricard-Blum S. The Collagen Family. Cold Spring Harb Perspect Biol. 2011;3:a004978 10.1101/cshperspect.a004978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muratori M, Luconi M, Marchiani S, Forti G, Baldi E. Molecular markers of human sperm functions. Int J Androl. 2008;32:25–45. 10.1111/j.1365-2605.2008.00875.x [DOI] [PubMed] [Google Scholar]
- 98.Coulombe PA, Omary MB. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. [DOI] [PubMed] [Google Scholar]
- 99.Gu LH, Coulombe PA. Keratin function in skin epithelia: A broadening palette with surprising shades. Curr Opin Cell Biol. 2007;19:13–23. 10.1016/j.ceb.2006.12.007 [DOI] [PubMed] [Google Scholar]
- 100.Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313:2021–2032. 10.1016/j.yexcr.2007.03.005 [DOI] [PubMed] [Google Scholar]
- 101.Schäcke H, Schumann H, Hammami-Hauasli N, Raghunath M, Bruckner-Tuderman L. Two Forms of Collagen XVII in Keratinocytes a full-length transmembrane protein and a soluble ectodomain. J Biol Chem. 1998;273:25937–25943. [DOI] [PubMed] [Google Scholar]
- 102.Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, et al. Desmoplakin Is Required Early in Development for Assembly of Desmosomes and Cytoskeletal Linkage. J Cell Biol. 1998;143:2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fontao L, Favre B, Riou S, Geerts D, Jaunin F, Saurat JH, et al. Interaction of the Bullous Pemphigoid Antigen 1 (BP230) and Desmoplakin with Intermediate Filaments Is Mediated by Distinct Sequences within Their COOH Terminus. Mol Biol Cell. 2003;14:1978–1992. 10.1091/mbc.E02-08-0548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Martins ML, Onaka EM, Moraes FR, Bozzo FR, Paiva AMFC, Gonçalves A. Recent studies on parasitic infections of freshwater cultivated fish in the state of São Paulo, Brazil. Acta Sci Biol Sci. 2002;24:981985. [Google Scholar]
- 105.Martins ML, Souza VN, Moraes JR, Moraes FR, Costa AJ. Comparative evaluation of the susceptibility of cultivated fishes to the natural infection with myxosporean parasites and tissue changes in the host. Braz J Biol. 1999;59:263–269. [Google Scholar]
- 106.Tavares-Dias M, Neves LR, Santos EF, Dias MKR, Marinho RGB, Ono EA. Perulernaea gamitanae (Copepoda: Lernaeidae) parasitizing tambaqui (Colossoma macropomum) (Characidae) and the hybrids tambacu and tambatinga, cultured in northern Brazil. Arq Bras Med Vet Zootec. 2011;63:988–995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
Comparative distribution of the GO terms and the numbers of transcripts involved in Biological Processes (BP), Molecular Function (MF), and Cell Components (CC) in the C. macropomum and tambacu.
(TIF)
(TIF)
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(CSV)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Raw reads data that support the findings of this study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (Bioproject: PRJNA353388/ Acession number: SRX2375124, SRX2375122, SRX2375120, and SRX2375119).