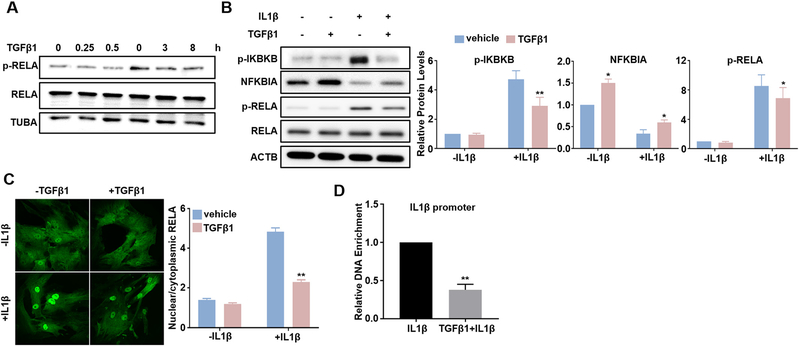
Fig. 5.
TGFβ1 suppresses IL1β-activated RELA/NF-κB pathway. Subconfluent HCASMCs were starved overnight, followed by TGFβ1 (2 ng/ml) treatment for the indicated time periods. Whole cell lysates were collected for western blot of RELA and p-RELA (A). Subconfluent HCASMCs were starved for 24 h, treated with TGFβ1 (2 ng/ml) overnight and then IL1β (4 ng/ml) for 15 min. Whole cell lysates were collected for western blot of the indicated NF-κB signal proteins and the related quantifications (B); immunofluorescence staining of RELA (green) (C); Chromatin Immunoprecipitation (ChIP) assay for RELA binding to IL1β promoter encompassing a predicted NF-kB site, amplified DNA signal was normalized to isoform and species matched IgG negative control (D). Representative western blot and fluorescence images from at least 3 independent experiments are shown. TUBA/ACTB is used as internal loading control. Quantitative data are expressed as means ± S.E.M. of at least 3 independent experiments, reflecting fold changes of TGFβ1 and/or IL1β treated to vehicle control values (set to 1; for D, data reflect fold changes of (TGFβ1 + IL1β) treated to IL1β treated values (set to 1)). *P < 0.05, **P < 0.01 compared to vehicle control, two-way ANOVA with Sidak’s post hoc test for multiple comparison correction (B, C); **p < 0.01 compared to IL1β treated group, unpaired two-tailed Student’s t-test (D). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)