Fig. 4.
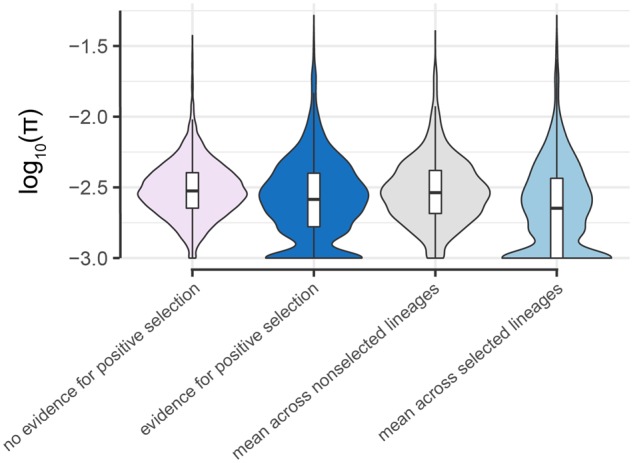
Nucleotide diversity near CpG sites is lower in lineages where positive selection has been inferred. Log10(π) (with an offset of 0.001 to avoid undefined values) for the 1-kb window surrounding CpG sites where DNA methylation levels are taxonomically structured, and 1) we inferred no evidence for positive selection (pink: n = 16,907 sites) or 2) where positive selection was inferred on any baboon lineage (dark blue: n = 3,043 sites, based on the intersection set of the heuristic and OU approaches for multispecies lineages and results from the heuristic approach for single lineages). Log10(π) for sites in dark blue are replotted in gray for lineages unaffected by putative positive selection and in light blue for lineages putatively affected by positive selection. Lineage-site combinations linked to positive selection (light blue) exhibit lower local nucleotide diversity than all other classes (Tukey’s Honestly Significant Difference test: P = 1.75 × 10−8 compared with sites with no evidence of positive selection [pink]; P = 0.0160 against the same sites, but with π averaged across all lineages [dark blue]; P = 5.00 × 10−9 against the same sites, but with π averaged across nonselected lineages only [gray]). π was calculated separately for each species prior to averaging, and is log transformed here for visualization purposes only. Box plots show median (black bar) and interquartile range (whiskers).
