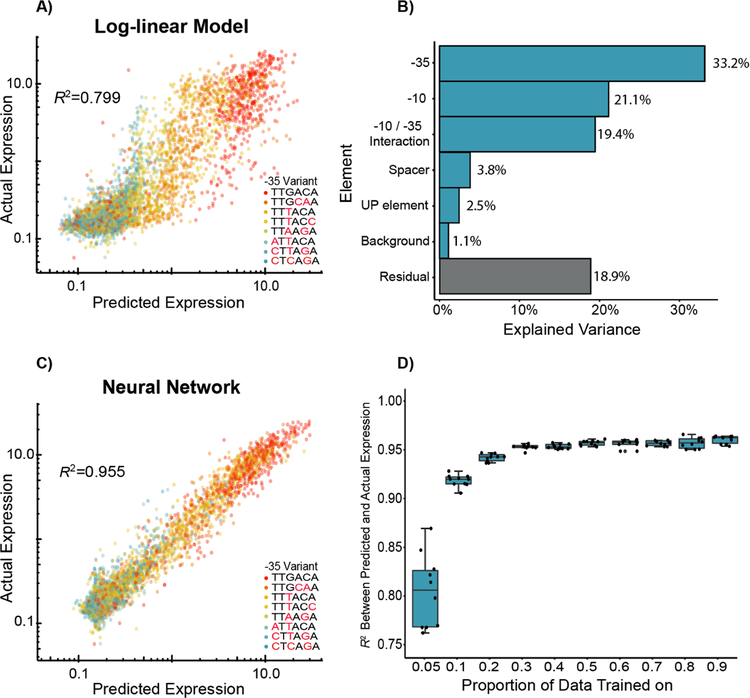
Figure 4. Predictive modeling of σ70 promoter strength.
A) We trained a log-linear model on 50% of the data, and the resultant predictions on the remaining data explain approximately 80% of the variance in expression within our dataset. B) We analyzed the model by ANOVA and found that approximately 73.7% of variance in promoter expression can be explained by the −10 and −35 elements (and their interaction). C) We also trained a simple neural network model and found that the resultant predictions captured an estimated 95.5% of the promoter variance, indicating that these models are better able to capture more complex interactions between sequence elements. D) We trained the same neural network models with 10-fold cross-validation and show that we can effectively predict promoter expression when trained on as little as 5% of the data. In 4A, 4C, and 4D, R2 is the coefficient of determination between predicted and actual expression values on the held-out datasets.