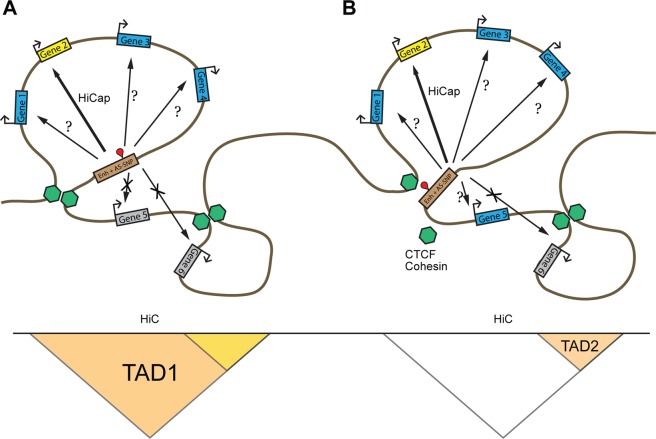
Figure 4.
Schematic representation of using 3D interactions data to prioritize candidate target genes. (A) AS-SNP (red pin) in a regulatory enhancer harbored in a TAD loop defined by HiC data. The genomic architecture reduces the pool of possible target genes to genes enclosed in the TAD (genes 1–4). HiCap analyses allow narrowing the list of putative target genes even further evaluating specific probe interactions, pointing in this example to gene 2 as the likely target for the enhancer element. (B) The presence of an AS-SNP in a TAD interaction domain may lead to an altered assembly of the TAD formation protein complex (e.g. CTCF, cohesin, etc. represented in green) resulting in a different TAD architecture. In this example the disruption of TAD1 extends the list of putative target genes to genes 1–5.