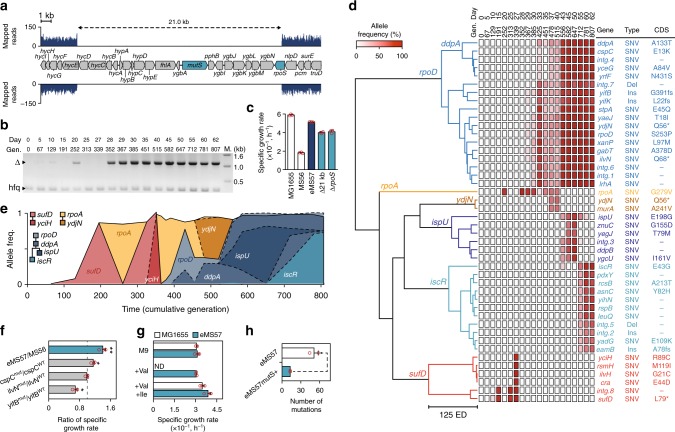
Fig. 2.
Whole-genome resequencing of adaptive laboratory evolution (ALE) experiment. a Spontaneous large deletion in eMS57 spanning 21 kb including 21 genes. No sequencing read was mapped onto this region. b The occurrence of large deletions was tracked by PCR. Δ indicates PCR amplicon only amplified from the deletion. hfq was used as a positive control of PCR. Gen and M indicate cumulative generations and a DNA marker, respectively. c Growth rates of MS56 with rpoS deletion or a large deletion compared to MG1655 and eMS57. Error bars indicate the s.d. of three individual cultures shown in red circles. d Heatmap indicates frequencies of mutations in a given population. Shown are 31 mutations with allele frequency >0.5 at least once during ALE. Dendrogram shows clonal lineages determined by hierarchical clustering. Gen: cumulative generations, SNV: single-nucleotide variation, *: stop codon, fs: frame shift, ED: Euclidean distance. e Three lineages and their sub-lineages were inferred from hierarchical clustering. yciH sub-lineage emerged within the sufD lineage. ddpA, ispU, and iscR are sub-lineages of the rpoD lineage. ydjN is a sub-lineage of the rpoA lineage. Sub-lineages are presented as dotted lines. f Construction of the cspC point mutation on MS56 had a beneficial effect on fitness, whereas ilvN mutation showed no effect. The yifB mutation on MS56 decreased the growth rate. Error bars indicate s.d. of three biological replicates shown as red circles. *P value <0.05, **P value <0.01 (two-sided Welch’s t test, a difference between growth rate of wild-type and mutant were tested, n = 3). g Growth rate of MG1655 and eMS57 in response to valine supplementation which inhibits cell growth (white bar: MG1655, blue bar: eMS57, error bars indicate s.d. of three biological replicates shown as red circles). eMS57 was completely resistant to valine toxicity. Addition of isoleucine compensated for the inhibitory effect of valine in MG1655. h Additional 300 ALE generations of eMS57 and eMS57mutS+ revealed that inactivation of mutS increased the mutation rate of eMS57. Error bars indicate s.d. of two independent ALE populations (shown as red circles). Source data are provided as a Source Data file