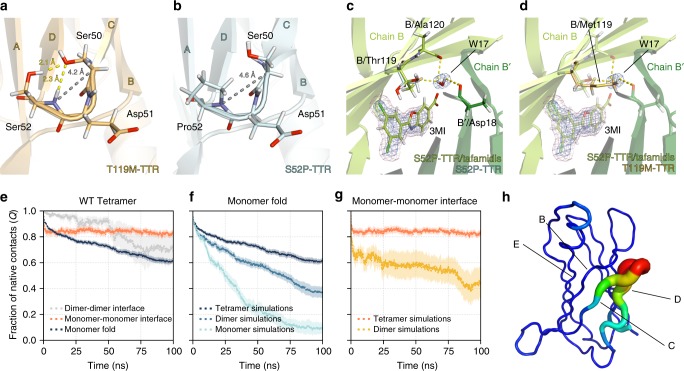
Fig. 3.
Neutron structures and molecular dynamics results. a In the T119M mutant (as well as in the WT; not shown), the Ser52 residue forms two hydrogen bonds (yellow dash lines) with the Ser50 residue. The distance between Ser50-Cα and the amide-N of Ser52 is 4.2 Å (grey), and it is the same in the WT (not shown). b In the S52P mutant, due to the absence of the hydrogen bonds between Pro52 and Ser50, the distances between Ser50-Cα and the amide-N of Pro52 is longer (4.6 Å), creating a looser CD loop. c Binding of tafamidis (3MI) results in a change of orientation for residue Thr119 (cyan, before binding; green, after binding). In the S52P/tafamidis complex, the hydroxyl side chain of Thr119 forms a hydrogen bond to a water molecule. d The water molecule in the S52P/tafamidis complex occupies the same position as the side chain of Met119 residue (light orange) in the non-pathogenic T119M mutant. The binding site between chain B and B′ is shown. The blue and magenta mesh show the 2Fo-Fc neutron scattering length density map and the 2Fo-Fc X-ray electron density map, respectively. Maps were contoured at 1.5 σ. e–g Loss of native fold and contacts in WT-TTR during high-temperature MD simulations with the Amber99sb*-ILDNP force field; shown are the mean and its standard error from 10 simulation repeats (results for Charmm36 are in Supplementary Figure 6). e Degree of disruption of the monomer fold, the monomer–monomer interface, and the dimer–dimer interface, when simulating the WT tetramer over the course of 100 ns. f Degree of unfolding of TTR monomers as part of a tetramer, dimer, or monomer in solution. g Degree of disruption of the monomer–monomer interface when simulating the dimer and the tetramer. h Protein regions in the PLS-FMA mode contributing most to the change in the fraction of native contacts for the high-temperature simulations