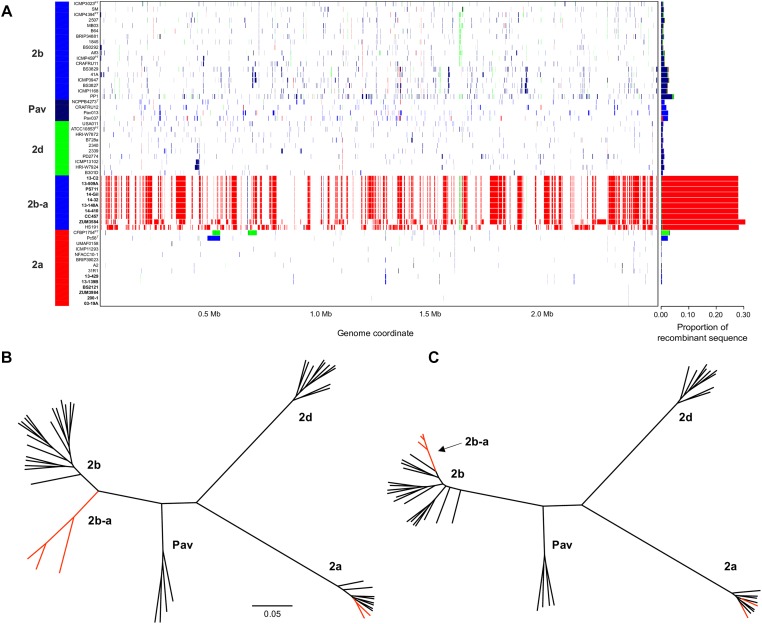
FIGURE 2.
Inference of recent homologous recombination between P. syringae sensu stricto phylogroups. Distribution, origin, and proportion of recent recombination events across the P. syringae core genome as predicted by fastGEAR. The lineage predictions are color coded and phylogroups labeled accordingly. Strains isolated from cucurbits hosts are shown in bold (A). Unrooted, maximum likelihood phylogeny based on 246,510 core genome SNPs (B) and unrooted, maximum likelihood phylogeny based on 81,421 recombination free SNPs (C). Branches corresponding to strains isolated from cucurbit hosts and the millet strain, HS191, are labeled in red. The scale bar indicates the number of substitutions per site.