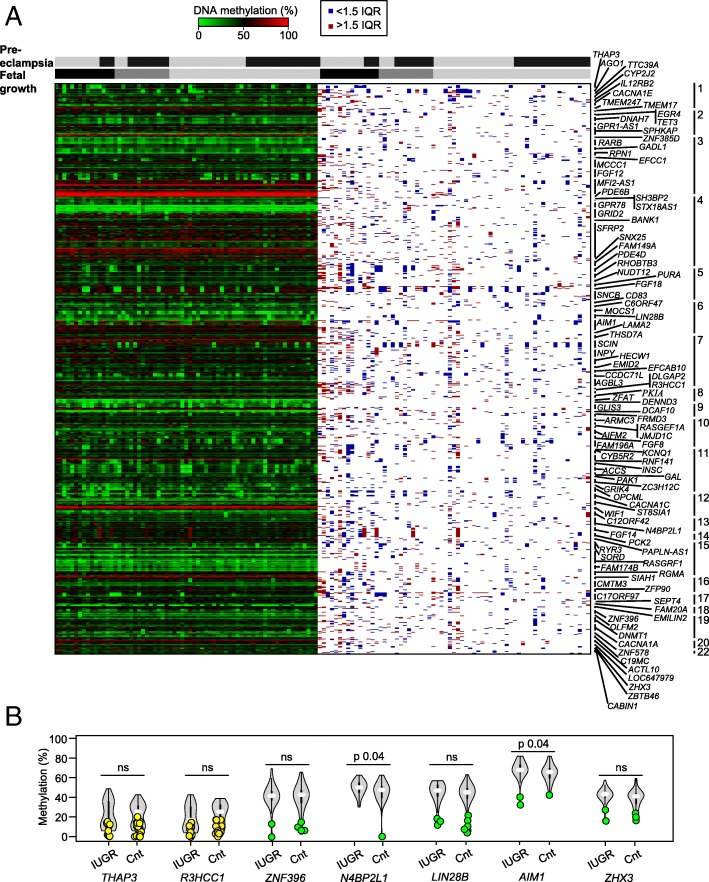
Fig. 2.
Analysis of imprinted methylation at placenta-specific DMRs using HM450k methylation arrays. a The left side is a heatmap of absolute methylation (β values) for individual Infinium probes mapping to 112 DMRs in 67 placenta samples. The right side of the figure reveals outlier samples as identified by > 1.5 IQR (red) and < 1.5 IQR (blue) methylation values, respectively. Samples are classified by both the presence of pre-eclampsia (black yes; gray no) and fetal growth parameters (appropriate for gestational age light gray; SGA dark gray; IUGR black) b Pyrosequencing confirmation of the aberrant methylation profiles identified using HM450k arrays, as well as the quantification of the extended placenta cohort. The violin plots include the median (white dot) and the interquartile range (black rectangle); samples with hypomethylation defined by < 1.5 IQR are highlighted as green data points. Placenta-specific DMRs with a distinct population of lowly methylated samples not identified by IQR statistic are highlighted in yellow. The non-parametric Mann-Whitney-Wilcoxon test was used to calculate the significance of the differences between IUGR and control groups (ns indicated no significance, p > 0.05)