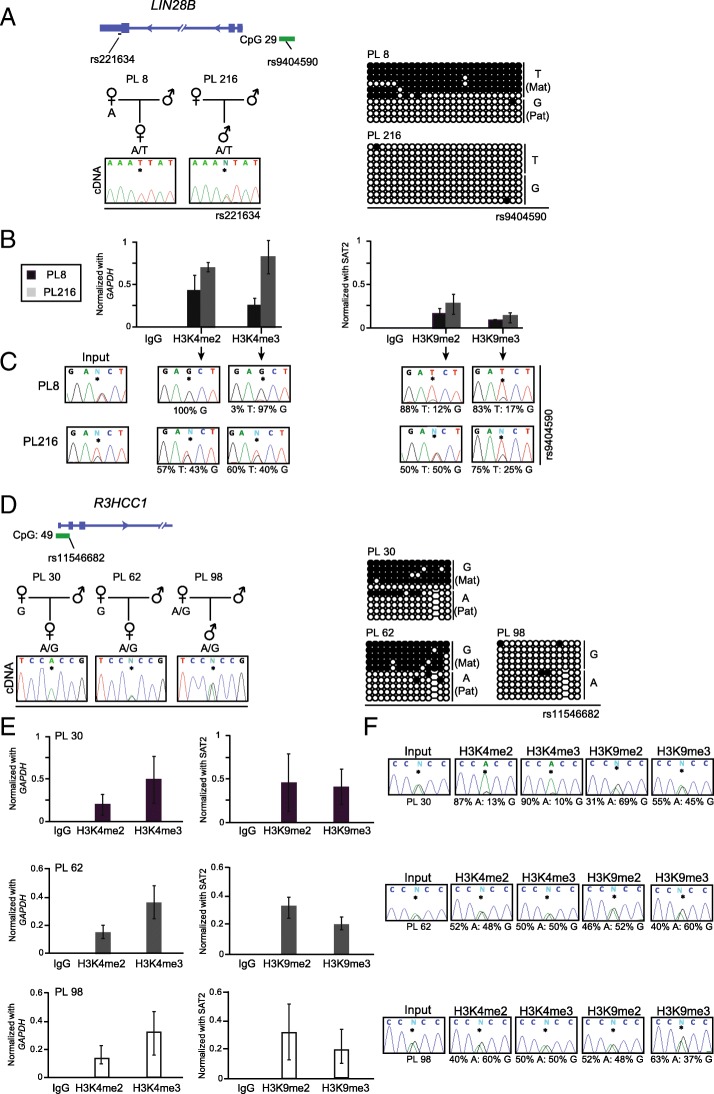
Fig. 3.
Detailed characterization of histone modifications within placenta-specific DMRs in samples with polymorphic imprinting. Schematic representation of a the LIN28B loci, indicating the position of transcripts and CpG islands incorporating the DMRs. The methylation of the two placenta samples analyzed was assessed by bisulphite PCR and sub-cloning. Each circle represents a single CpG on a DNA strand. (•) methylated cytosine, (o) unmethylated cytosine. Each row corresponds to an individual cloned sequence with the genotype indicated for the heterozygous SNP incorporated into the amplicon. b Quantitative PCR targeting the LIN28B DMR in ChIP material. Precipitations were normalised to GAPDH promoter (H3K4me2/3) and SAT2 repeats (H3K9me2/3). The graphs represent the mean values (± standard deviation). For each placenta sample, values are the mean of at least three independent ChIP experiments, each in triplicate. c The allelic distribution of each mark was determined by direct sequencing of the PCR product encompassing the heterozygous SNP used for the methylation analysis. The percentage of allelic enrichment is shown under the electropherograms. d Diagram of the R3HCC1 loci and the methylation bisulphite PCR profiles of the three analyzed samples. e, f The quantitative and allelic ChIP results for the R3HCC1 DMR, respectively, which were analyzed in the same way as for LIN28B