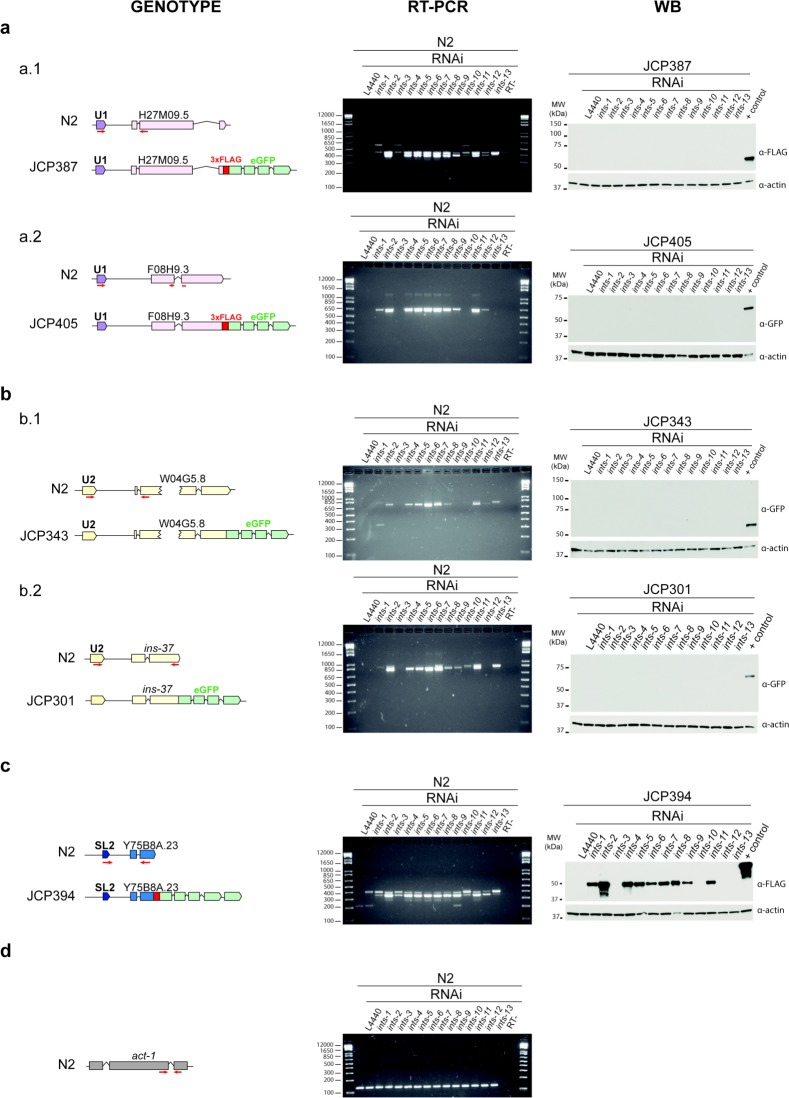
Fig 4. Knockdown of C. elegans integrator subunits results in formation of long “chimeric sn-mRNAs” that can be translated to proteins in the case of “chimeric SL-mRNAs”.
Left panel: schematic representation of the snRNA genes analysed. Red arrows represent the primer pairs used. Middle panel: gel electrophoresis showing RT-PCR analysis. RT minus controls do not show genomic DNA contamination. Double bands correspond to the spliced (lower band) and unspliced (upper band) mRNA fractions. Right panel: Western blot to check the translation of “chimeric sn-mRNAs” into proteins. (a) U1 type sn-mRNAs. a.1 PCR expected length: 396 bp (genomic: 439 bp). H27M09.5 estimated molecular weight: 68.8kDa. a.2 PCR expected length: 637 bp. F08H9.3 estimated molecular weight: 46.6 kDa. The U1 derived sn-mRNAs were not translated into proteins (b) U2 type snRNAs. b.1 PCR expected length: 782 bp (genomic: 840 bp). W04G5.8 estimated molecular weight: 72.7 kDa b.2 PCR expected length: 804 bp (genomic: 856 bp) INS-37 estimated molecular weight: 45.6 kDa. The U2 derived sn-mRNAs were not translated into proteins (c) SL-2 snRNA. PCR expected length: 358 bp (genomic 410bp). Y75B8A.23 estimated molecular weight: 39.5 kDa. The SL derived sn-mRNAs were translated into proteins. (d) Actin was used as the loading control: 41.8 kDa. PCR expected length: 156 bp (genomic: 208 bp).