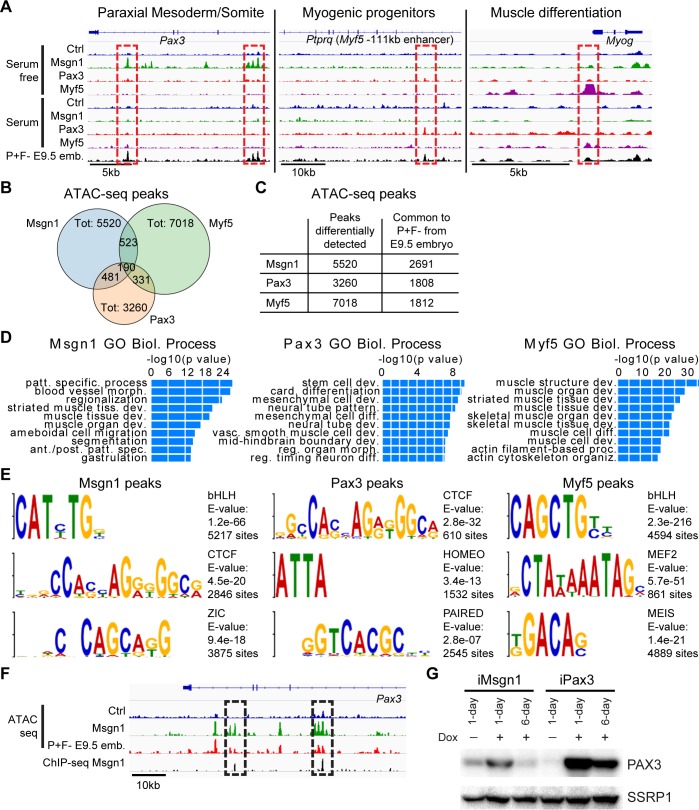
Fig 2. Pax3 as a gatekeeper of the myogenic lineage.
(A) Representative IGV tracks for genes associated with paraxial mesoderm/somite formation, myogenic progenitor specification (“spec.”), and muscle differentiation (“diff.”) and comparison with PDGFRα+FLK1− cells isolated from E9.5 mouse embryos (“P+F- E9.5 emb.”). (B) Number of ATAC-seq peaks from serum-free differentiation identified upon induction of Msgn1, Pax3, and Myf5. Venn diagram indicates the overlap between these 3 datasets. (C) Number of peaks from serum-free differentiation overlapping to ATAC-seq peaks detected in PDGFRα+FLK1− cells isolated from E9.5 mouse embryos. (D) Functional classification of Msgn1-, Pax3-, and Myf5-induced accessible peaks from serum-free differentiation using GREAT. Complete annotation data are reported in S2 Table. (E) Selected motifs associated with Msgn1-, Pax3-, and Myf5-induced peaks in serum-free differentiation. (F) IGV snapshot of the Pax3 locus. Changes in chromatin accessibility induced by Msgn1 in serum-free differentiation overlap to peaks identified in the Msgn1 ChIP-seq (dashed black squares). (G) Western blot analysis of PAX3 expression in Msgn1- and Pax3-induced cultures following 1-day and 6-day dox treatment. SSRP1 was used as loading control (“Ctrl”). Numerical values are available in S1 Data. ant./post., anterior–posterior; ATAC-seq, assay for transposase-accessible chromatin sequencing; bHLH, basic helix-loop-helix; Biol., biological; card., cardiac; ChIP-seq, chromatin immunoprecipitation sequencing; CTCF, CCCTC-binding factor; dev., development; dox, doxycycline; E, embryonic day; GO, gene ontology; IGV, Integrative Genomics Viewer; MEF2, myocyte enhancer factor 2; morph., morphology; Msgn1, mesogenin 1; Myf5, myogenic factor 5; organiz., organization; patt., pattern; Pax3, paired box 3; proc., process; reg., regulation; SSRP1, structure-specific recognition protein; Tot, total; vasc., vascular.