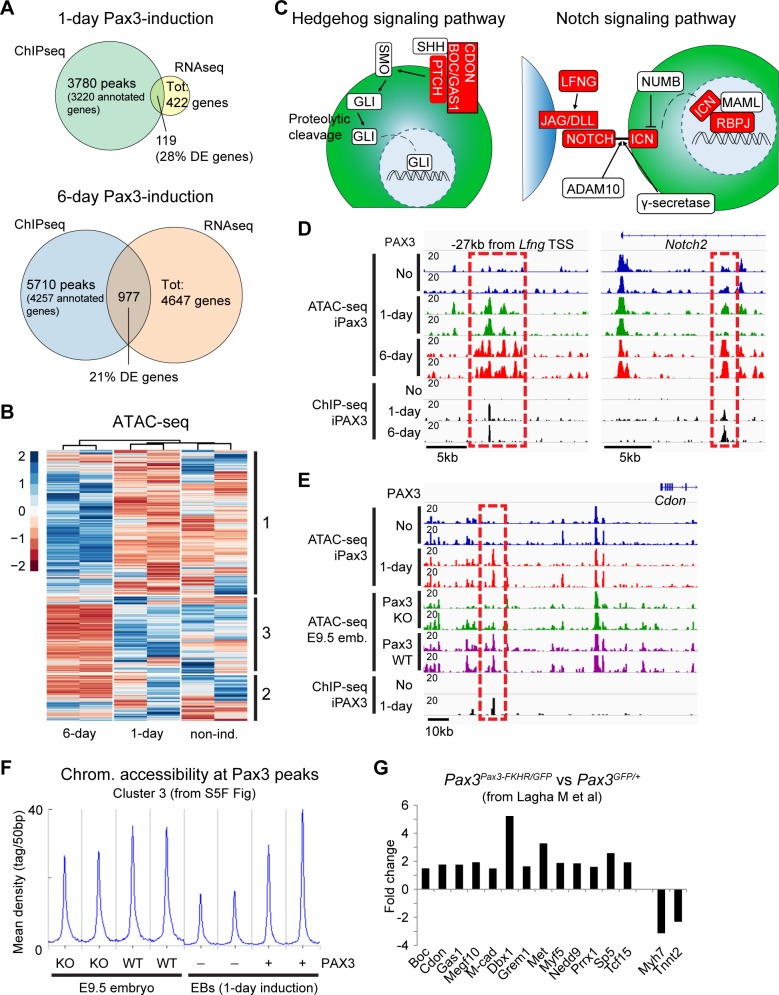
Fig 4. Pax3 induces chromatin remodeling at a subset of bound loci in mesodermal cells.
(A) Venn diagrams displaying overlap between DE genes and Pax3-bound loci in 1-day (top) and 6-day (down) samples. Numbers indicate the overlap between the two datasets. (B) Heatmap displaying the changes in chromatin accessibility in PDGFRα+FLK1− cells isolated from noninduced (“non-ind.”), 1-day, and 6-day Pax3-induced cells. Differential accessible loci from 1-day versus noninduced and 6-day versus noninduced were combined in a list of unique peaks and used to generate the differential analysis. Three clusters (indicated on the right side) were identified, and the corresponding coordinates were used for GO analysis. The legend indicates the scaled (z score) coverage information for each region. (C) Schematic representation of Hedgehog and Notch signaling pathways. Red blocks indicate genes characterized by increased chromatin accessibility upon Pax3 induction. (D) IGV track displaying chromatin accessibility and Pax3 genomic binding at the Lfng and Notch2 loci in cells isolated from 1-day and 6-day Pax3-induced (+) and noninduced (-) EB cultures. Dashed red squares show Pax3-dependent regulation of chromatin accessibility at the Pax3-bound Lfng −25 kb and Notch2 +10 kb sites. Pax3 binding to these elements are shown by the ChIP-seq tracks. (E) IGV track displaying chromatin accessibility and Pax3 genomic binding at the Cdon locus in cells isolated from 1-day Pax3-induced (+) and noninduced (-) EB cultures and Pax3 WT and KO E9.5 embryos (“emb.”). Dashed red square shows Pax3-dependent regulation of chromatin accessibility during embryonic development at the Cdon −49 kb site. (F) Distribution of ATAC-seq reads across a subset of 684 Pax3 ChIP-seq peaks generated using k-means clustering. Curves show chromatin (“chrom.”) accessibility at Pax3 peaks (±3 kb from peak center) in PDGFRα+FLK1− cells from Pax3 WT and KO embryos and noninduced (-) and 1-day induced (+) EB cells. (G) Selected Pax3 targets are regulated during mouse embryonic development (transcriptomic data published by [43]). Graph represents the expression fold change of selected statistically significant and differentially regulated transcripts from the somites of Pax3Pax3FKHR/GFP and Pax3GFP/+ embryos. Numerical values are available in S1 Data. ADAM, a disintegrin and metallopeptidase domain 10; ATAC-seq, assay for transposase-accessible chromatin sequencing; BOC, biregional Cdon binding protein; CDON, cell adhesion molecule–related/down-regulated by oncogenes; ChIP-seq, chromatin immunoprecipitation sequencing; DE, differentially expressed; DLL, delta-like canonical Notch ligand; E, embryonic day; EB, embryoid body; GAS1, growth arrest specific 1; GO, gene ontology; ICN, intracellular notch; IGV, Integrative Genomics Viewer; JAG, jagged; KO, knockout; LFNG, lunatic fringe; MAML, Mastermind-like; Pax3, paired box 3; PTCH, patched; RNAseq, RNA sequencing; SHH, Sonic hedgehog; SMO, Smoothened; Tot, total; TSS, transcription start site; WT, wild-type.