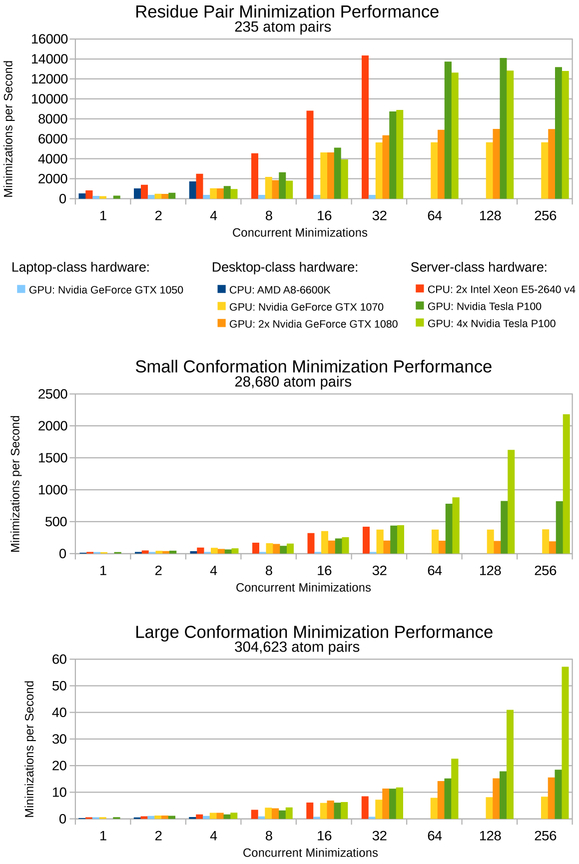
Figure 3: Benchmarks for protein conformation minimization in osprey 3.0 for various hardware platforms and for conformations of varying size.
From smallest to largest: (top) a single residue pair is the smallest multi-body minimization possible, (middle) a full protein conformation with a single flexible residue represents a small design, (bottom) a full protein conformation with 20 flexible residues represents a large design. For CPU hardware, concurrent minimizations correspond to CPU threads. For GPU hardware, concurrent minimizations correspond to streams defined by the CUDA framework. Faster minimization speeds correspond with faster osprey runtimes. All minimizations were performed on the Atx1 metallochaperone protein (PDB ID: 1CC8)47. Flexible residues were modeled with continuous sidechain flexibility, and all other residues remained completely fixed.