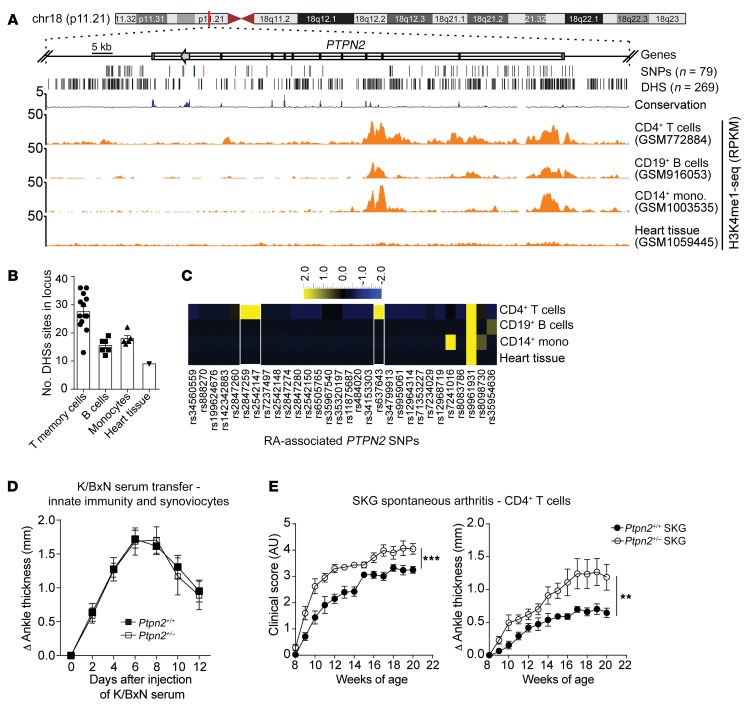
Figure 1. RA-associated haploinsufficiency of Ptpn2 promotes T cell–dependent arthritis in mice.
(A) UCSC tracks showing the chromosomal location of the human PTPN2 gene, containing a large haplotype block of RA-associated SNPs. Black lines indicate SNPs’ genomic location (the characterizing SNPs rs2847297, rs1893217, and rs8083786 are indicated in red), and DNase hypersensitivity sites (DHSs). Example tracks of H3K4me1-seq from CD4+ T cells, CD19+ B cells, CD14+ monocytes, and heart tissue. RPKM, reads per kilobase of transcript per million mapped reads. (B) Number of DHSs in the PTPN2 locus in single data sets of 4 primary cell types. (C) Heatmap of RA-associated SNPs (columns) that overlap with DHSs in different primary cell types (rows). (D) Development of K/BxN serum-induced arthritis in Ptpn2+/+ (n = 9) and Ptpn2+/– (n = 7) male BALB/c mice. (E) Clinical score and ankle thickness during development of spontaneous arthritis in female Ptpn2+/+ (n = 8) and Ptpn2+/– (n = 8) SKG mice. Compiled data from at least 2 independent experiments are shown in D and E. Arthritis severity was quantified using the area under the curve. Bars represent mean ± SEM. **P < 0.01, ***P < 0.001 by Mann-Whitney.