Figure 2.
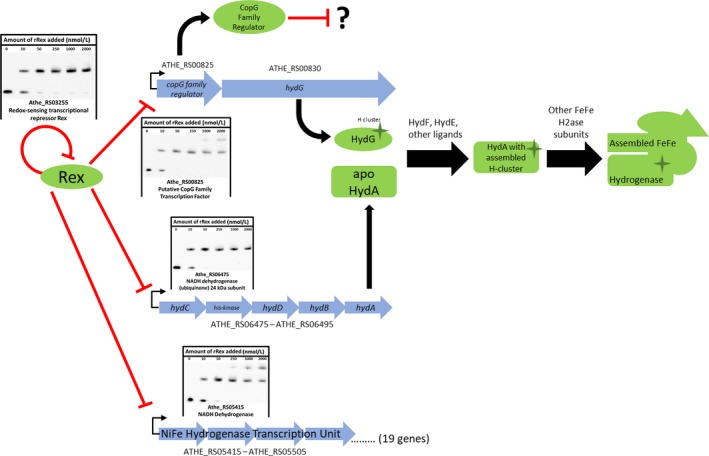
Proposed models of Rex repression supported by electromobility shift assays of Rex‐binding sites upstream of hydrogenase genes in C. bescii. These results suggest Rex represses FeFe hydrogenase structural genes and hydG, a maturase necessary for active site assembly in FeFe hydrogenases. Rex represses expression of NiFe hydrogenase structural genes. Rex also autoregulates itself. Genomic arrangement of hyd and ech genes inferred from homology to putative transcriptional units identified in C. saccharolyticus (Van de Werken et al., 2008) and predicted transcriptional units identified, using the DOOR prokaryote operon database (Mao et al., 2014). Hyd subunit assembly scheme adapted from (Kuchenreuther et al., 2014)
