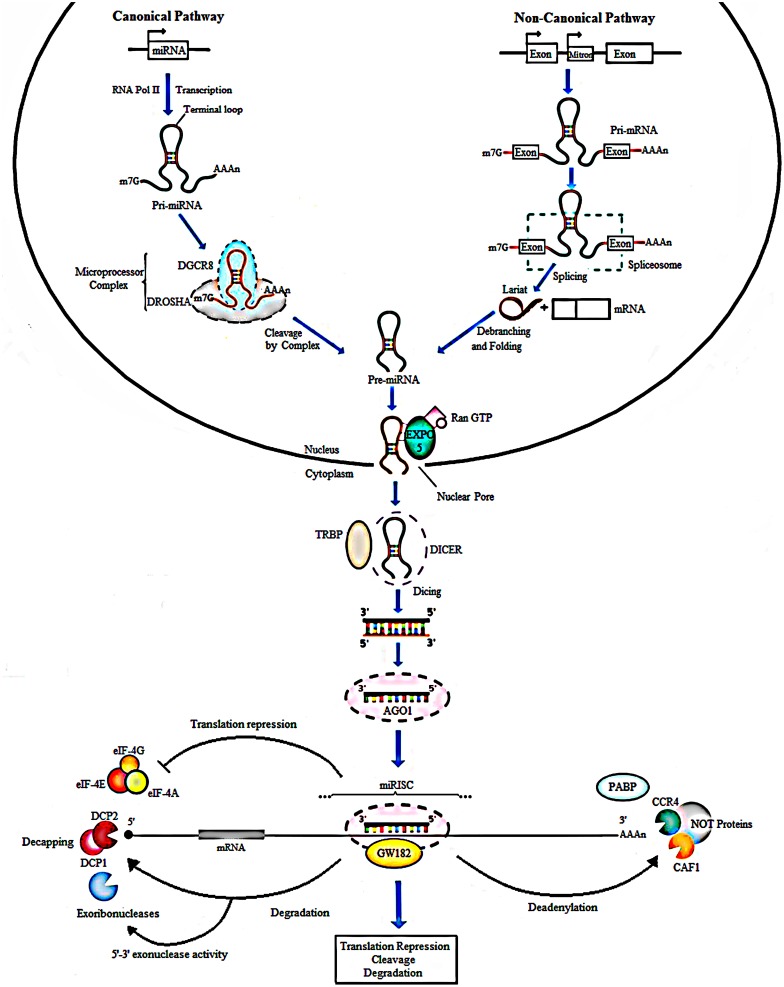
FIGURE 1.
Biogenesis of miRNAs. In the canonical pathway, miRNAs are transcribed from their loci by RNA polymerase II into a long primary transcript of about 80 nucleotides called the pri-miRNA. Cleavage follows and is done by Drosha, a type III RNase along with the DGCR8 protein to produce pre-miRNA. In the non-canonical pathway, Mirtrons are spliced by the spliceosome to form looped intermediates referred to as lariat which then refold into pre-miRNAs. Next, the exportin 5, a RAN-GTP dependent transporter, mediates the movement of pre-miRNAs from the nucleus into the cytoplasm. Further processing by Dicer and TARBP2 protein generates mature miRNAs, producing double-stranded structure of miRNA of about 21-22 nt in length. The duplex is loaded into an AGO protein. The passenger strand (miRNA∗) is degraded, whereas the guide strand is incorporated by the Ago into the miRNA-induced silencing complex (miRISC). In animals, imperfect complementarity occurs when the miRNA seed region, nucleotides 2-8, BPs perfectly with the complementary seed match site in the 3′ UTR of the target mRNA resulting in translational repression or degradation. GW182 a core component of miRISC, mediates deadenylation of mRNAs by interacting with AGO and PABP consequently leading to recruitment of deadenylases like CCR4 and CAF1. Translation repression can result from inhibited binding of PABP to the poly (A) tail of the mRNA, responsible for attracting the elFs to mRNA to initiate translation. The formation of the CCR4-CAF1-NOT complex, a poly A tail-truncating enzyme, mediated by binding of miRISC to mRNA results in truncation of the downstream poly A tail, reduced binding of translation initiation factors and translation repression. The shortening or complete removal of the poly (A) tail induces the removal of the 5′ cap of the mRNA. Decapping is also mediated by DCP1 and DCP2. Consequently, the uncapped mRNA is rapidly degraded by 5′-3′ exoribonucleases.