Abstract
The human fungal pathogen Candida glabrata appears to utilise unique stealth, evasion and persistence strategies in subverting the onslaught of host immune response during systemic infection. However, macrophages actively deprive the intracellular fungal pathogen of glucose, and therefore alternative carbon sources probably support the growth and survival of engulfed C. glabrata. The present study aimed to investigate the role of the glyoxylate cycle gene ICL1 in alternative carbon utilisation and its importance for the virulence of C. glabrata. The data showed that disruption of ICL1 rendered C. glabrata unable to utilise acetate, ethanol or oleic acid. In addition, C. glabrata icl1∆ cells displayed significantly reduced biofilm growth in the presence of several alternative carbon sources. It was also found that ICL1 is crucial for the survival of C. glabrata in response to macrophage engulfment. Disruption of ICL1 also conferred a severe attenuation in the virulence of C. glabrata in the mouse model of invasive candidiasis. In conclusion, a functional glyoxylate cycle is essential for C. glabrata to utilise certain alternative carbon sources in vitro and to display full virulence in vivo. This reinforces the view that antifungal drugs that target fungal Icl1 have potential for future therapeutic intervention.
Introduction
Invasive candidiasis is a potentially lethal fungal infection caused by fungi from the Candida genus that is associated with high morbidity and mortality. Life-threatening blood stream infections (candidaemia) and deep-seated candidiasis are commonly seen in critically ill individuals such as intensive care unit (ICU) patients with predisposing host factors or underlying malignant diseases1,2. Over the last decade, the proportion of invasive candidiases caused by the predominant species Candida albicans has decreased. Meanwhile, there has been a corresponding shift towards certain non-Candida albicans Candida (NCAC) species, probably due to the selection imposed by antifungal drugs2–4. Candida glabrata has emerged as one of the most prominent invasive candidiasis-causing species, particularly in some of the European countries, USA, Canada and Australia5.
Numerous studies have focussed on C. albicans, and less attention has been devoted to the pathogenic attributes of C. glabrata. In C. albicans, hypha-mediated penetration is crucial for the invasion of the host epithelial cells through protruding filaments and secretion of hydrolytic enzymes and candidalysin6,7. In addition, the aggressive nature of C. albicans leads to stronger pro-inflammatory cytokine responses in the host. In contrast, C, glabrata is a haploid, non-dimorphic fungus that is incapable of hypha formation, and this pathogen seems to favour a ‘stealth and concealment’ approach during infection to avoid direct confrontation with immune cells8. Despite the lower pathogenicity of C. glabrata in comparison to C. albicans, the high mortality rate associated with invasive candidiasis caused by C. glabrata would argue otherwise. Therefore, C. glabrata likely possesses potent pathogenic attributes that do not relate to phenotypic dimorphism.
Interestingly, C. glabrata has been shown to elicit a unique cytokine profile that promotes the recruitment of monocytes instead of neutrophils9. Since C. glabrata survives and replicates within the hostile microenvironment of macrophages, but not in neutrophils10,11, it is possible that C. glabrata exploits these immune cells to survive against the neutrophil onslaught during the establishment of an infection. Upon engulfment by macrophages, C. glabrata reprograms its metabolic activity in order to adapt to nutrient deprivation (e.g. carbon starvation). Roetzer et al.12 have reported that C. glabrata counteracts nutrient deprivation via mobilization of intracellular resources through autophagy12. Autophagy, particularly pexophagy, is an important virulence factor in C. glabrata that is crucial to sustain this pathogen during carbon starvation. In addition, Ng et al.13 have shown that SNF3, which encodes a high affinity glucose sensor, is also important for C. glabrata to thrive within macrophages - a microenvironment with limited glucose availability13. In addition to autophagy and enhanced glucose sensing, alternative carbon utilisation is believed to be important for the survival and pathogenicity of Candida species. Transcriptional analyses of C. albicans and C. glabrata revealed extensive metabolic reprogramming that reflects adaptation to nutrient deprivation following macrophages engulfment14,15. This reprogramming includes the upregulation of genes from three interconnected alternative carbon utilisation pathways: gluconeogenesis (FBP1 and PCK1), the glyoxylate cycle (ICL1 and MLS1) and fatty acid β-oxidation (FOX2 and POX1). Upregulation of these pathways indicates that the macrophage actively deprives C. albicans and C. glabrata of their preferable carbon source, thus forcing these fungal pathogens to tune their metabolism to alternative carbon sources.
The ability to utilise alternative carbon sources is important for C. glabrata in many host niches. For example, lactate assimilation is required for the survival in the intestine16. In addition, it has been shown that vaginal isolates of C. glabrata are able to utilise acetate, even in the presence of glucose17. The scavenging of alternative carbon sources, such as acetate, is dependent on a functional glyoxylate cycle18. The glyoxylate cycle bypasses the two decarboxylation steps in the tricarboxylic (TCA) cycle, thereby permitting the assimilation of this carbon. The glyoxylate cycle depends upon two enzymes, isocitrate lyase and malate synthase, to produce malate, an intermediate of the TCA cycle. The glyoxylate cycle is absent from mammalian tissues, but is conserved in protists, archaea, plants, bacteria, fungi and nematodes19. Muñoz-Elías & McKinney (2005) showed that disruption of the genes that encode ICL isoforms in bacterial pathogens, ICL1 and ICL2, impairs the growth and persistence of Mycobacterium tuberculosis in vivo20. Mutants lacking both icl1 and icl2 showed a defect in intracellular replication and were rapidly eliminated from the mice. Isocitrate lyase is also required for Salmonella enterica serovar Typhimurium during chronic infection and is essential for the virulence of Rhodococcus equi and Pseudomonas aeruginosa21–23.
With regard to medically important fungi, isocitrate lyase is required for the growth of Aspergillus fumigatus on alternative carbon sources such as acetate, ethanol and fatty acids24. Nevertheless, isocitrate lyase is not required for the establishment of invasive aspergillosis in murine model25. Similarly, although ICL1 is highly induced in the presence of alternative carbon sources and in rabbit meningitis model, ICL1 mutants of Cryptococcus neoformans show no apparent virulence defect in murine or rabbit infection models in vivo26. In C. albicans, disruption of the key glyoxylate cycle gene ICL1 severely attenuates virulence in murine models of invasive candidiasis27,28. These studies suggest that the degree to which the glyoxylate cycle contributes to human pathogenicity depends on the species of fungal pathogen. To date, the significance of the glyoxylate cycle in C. glabrata pathogenicity remains unknown. Therefore, taking cues from C. albicans, we have investigated the role of ICL1 in the metabolic flexibility and virulence of C. glabrata.
Results
ICL1 is essential for the growth of C. glabrata on certain alternative carbon sources
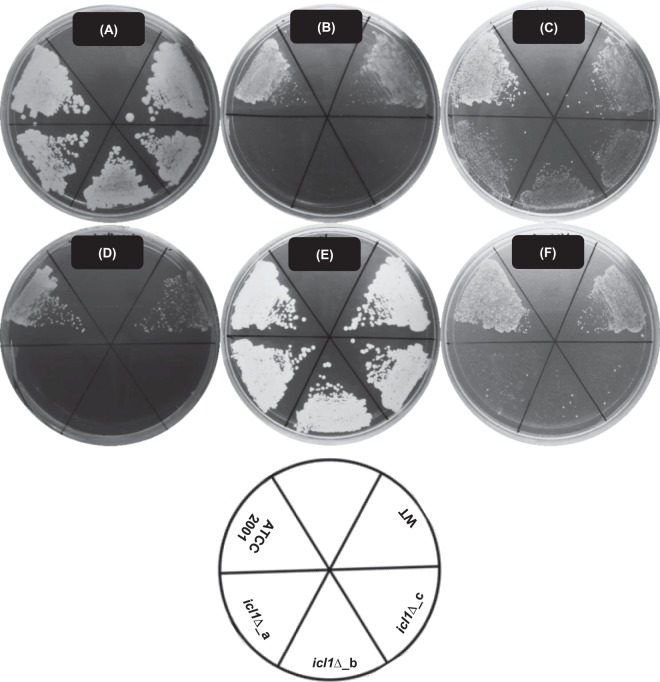
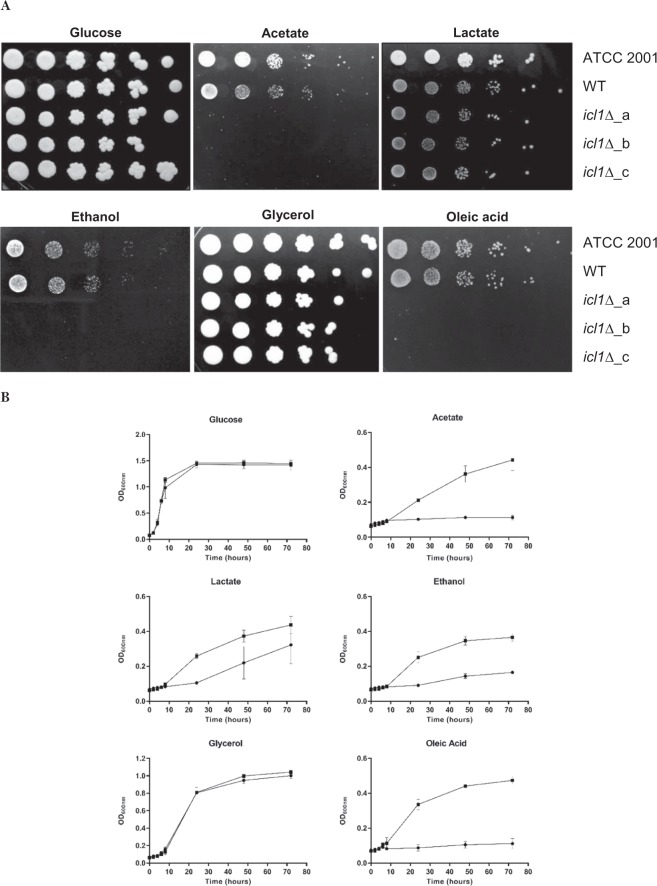
First, the ability of C. glabrata ATCC 2001, WT and icl1∆ cells to grow on glucose or alternative carbon sources was tested using simple growth assays. As anticipated, C. glabrata ATCC 2001 is able to utilise all of the alternative carbon sources tested, in addition to the preferred carbon source, glucose. The WT and mutant strains lacking ICL1 were all viable and they grew equally well in the presence of glucose or glycerol as sole carbon source. There was a slight decrease in the growth of mutant strains on lactate (Figs 1 and 2). We also found that ICL1 deletion rendered C. glabrata unable to grow on acetate and ethanol as sole carbon source (Figs 1 and 2). In addition, C. glabrata icl1∆ cells grew poorly in media containing oleic acid (Figs 1 and 2). Similar carbon utilisation profiles were obtained for three independently constructed C. glabrata icl1 mutants (icl1∆_a, icl1∆_b and icl1∆_c). We conclude that, in C. glabrata, ICL1 is indispensable for the utilisation of acetate, ethanol and oleic acid, and partially required for the utilisation of lactate.
Figure 1.
Representative image of growth phenotypes of C. glabrata ATCC 2001, WT and icl1∆ mutants (icl1∆_a, icl1∆_b and icl1∆_c) on glucose and alternative carbon sources. C. glabrata strains were grown on SC media containing (A) 2% glucose, (B) 2% acetate, (C) 2% lactate, (D) 2% ethanol, (E) 2% glycerol or (F) 0.2% oleic acid as the sole carbon source for 96 h at 37 °C. All experiments were performed in triplicate and each independent experiment was repeated three times.
Figure 2.
(A) Growth of C. glabrata ATCC 2001, WT and icl1∆ mutants in spot dilution assay. Deletion of ICL1 renders C. glabrata unable to utilise and grow in SC media supplemented with 2% acetate, 2% ethanol or 0.2% oleic acids as the sole carbon source. (B) Growth profile of C. glabrata WT (■) and icl1∆ mutants (●) in liquid SC media supplemented with 2% glucose, 2% acetate, 2% lactate, 2% ethanol, 2% glycerol or 0.2% oleic acid as the sole carbon source. All experiments were performed in triplicate and each independent experiment was repeated three times.
ICL1 is essential for the formation of C. glabrata biofilms in certain alternative carbon sources
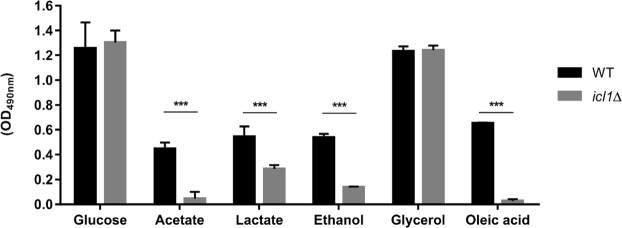
Since the deletion of ICL1 impacts the planktonic growth of C. glabrata on several alternative carbon sources, we then investigated the role of ICL1 in biofilm formation. To achieve this, we measured the metabolic activity of C. glabrata biofilms formed on different alternative carbon sources. As expected, icl1∆ cells displayed similar levels of biofilm formation to the WT control strain during growth on glucose (Fig. 3). In addition, ICL1 was not essential for biofilm formation in the presence of glycerol as the sole carbon source (Fig. 3). However, the disruption of ICL1 reduced C. glabrata biofilm formation on acetate, lactate, ethanol and oleic acid. Indeed, significant reductions in biofilm formation were observed for the icl1∆ cells on oleic acid (up to 95%: p < 0.001), closely followed by acetate (90%), ethanol (75%) and lactate (48%).
Figure 3.
Biofilm formation of C. glabrata WT and icl1∆ mutants in glucose and alternative carbon sources. Results are presented as means ± SD. *p < 0.05 was considered statistically significant relative to WT strain. All experiments were performed in triplicate and each independent experiment was repeated three times.
ICL1 is essential for the survival of C. glabrata cells following macrophage engulfment
Our results demonstrate that ICL1 is required for the metabolic flexibility of C. glabrata. Therefore, we reasoned that ICL1 might also play an essential role in promoting the survival of this fungus following phagocytosis by macrophages. RAW264.7 macrophages were challenged with C. glabrata and the survival of internalized fungal cells was determined by measuring the resultant colony forming units (CFUs). The results showed the icl1∆ mutant was much more susceptible to macrophage killing than the WT control strain (Fig. 4). This observation confirms the importance of ICL1 for the survival of C. glabrata following macrophage ingestion.
Figure 4.
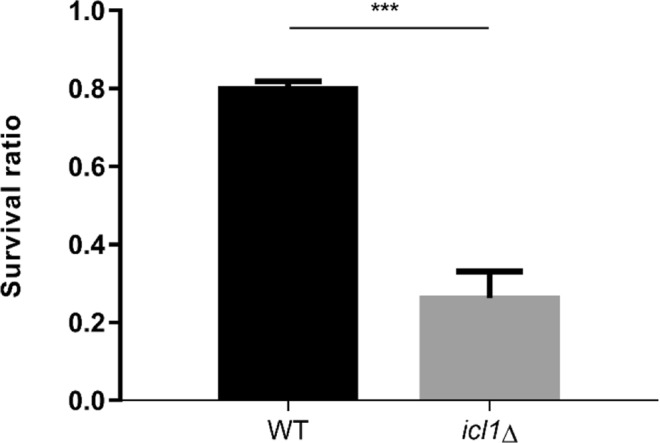
Survival ratio of internalised C. glabrata WT and icl1∆ mutants recovered from RAW264.7 macrophages. Results are presented as means ± SD. *p < 0.05 was considered statistically significant relative to WT strain. All experiments were performed in triplicate and each independent experiment was repeated three times.
ICL1 is essential for the virulence of C. glabrata in vivo
To investigate the relevance of ICL1 to the virulence of C. glabrata in vivo, the icl1∆ mutants were tested in a mouse model of invasive candidiasis. In this survival assay, equivalent doses of C. glabrata WT and icl1∆ (2 × 108 cells) were administered to immunocompromised Institute of Cancer Research (ICR) mice via lateral tail vein injection, and the mice were monitored for up to 21 days. Infection with the C. glabrata WT strain resulted in 50% mortality within the first three days and achieved 90% mortality at day 21 post-infection. In contrast, infection with the C. glabrata icl1∆ cells only resulted in 40% mortality (Fig. 5A), and the remaining mice survived up to 21 days post-infection. Mantel-Cox log rank analysis of survival curve demonstrated that the disruption of ICL1 confers a significant attenuation in the pathogenicity of C. glabrata in this murine model of invasive candidiasis (p < 0.05).
Figure 5.
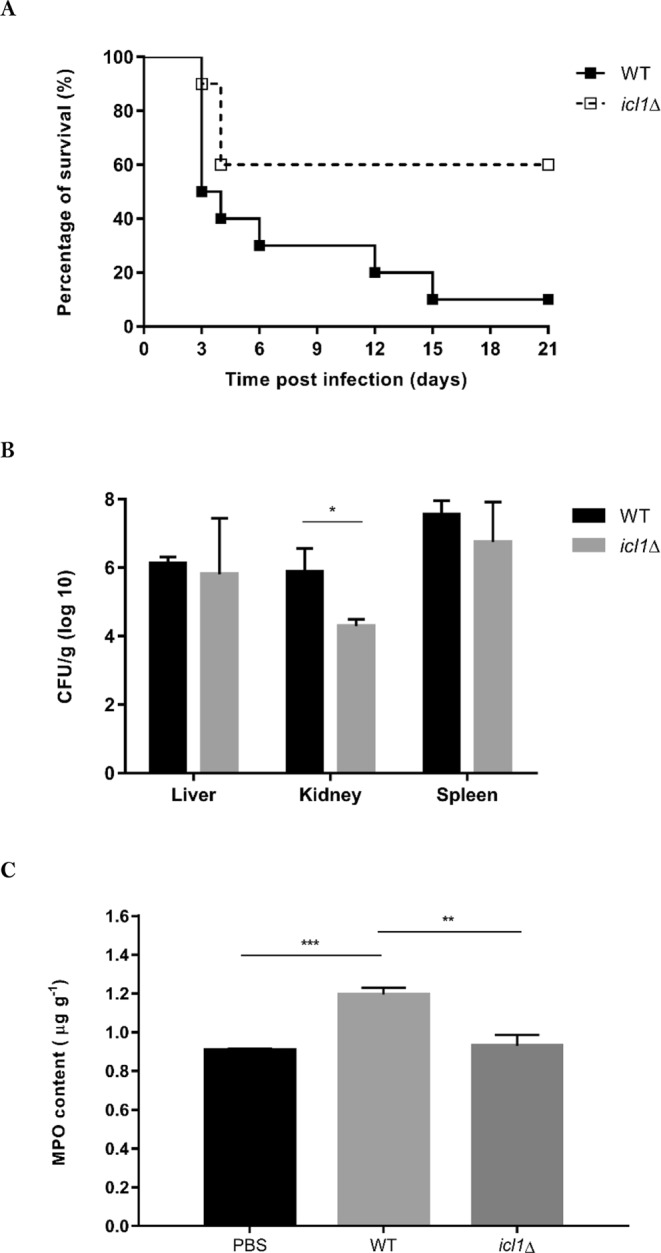
ICL1 is essential to maintain wild type level of C. glabrata virulence in vivo. (A) Survival curve of immunosuppressed ICR mice infected with C. glabrata WT (n = 10) or icl1∆ mutant (n = 10). Mantel-Cox log rank analysis of survival curve demonstrated that the virulence of icl1∆ mutant was significantly attenuated (p < 0.05) compared to WT strain. (B) Fungal burdens in different organs harvested from immunosuppressed ICR mice infected with C. glabrata WT or icl1∆ mutant. CFU counts were determined from cultures of tissue homogenates of five animals per group. (C) MPO content in kidney of immunosuppressed ICR mice infected with C. glabrata WT or icl1∆ mutant. MPO content were determined from cultures of kidney homogenates of five animals per group. *p < 0.05 was considered statistically significant relative to WT strain.
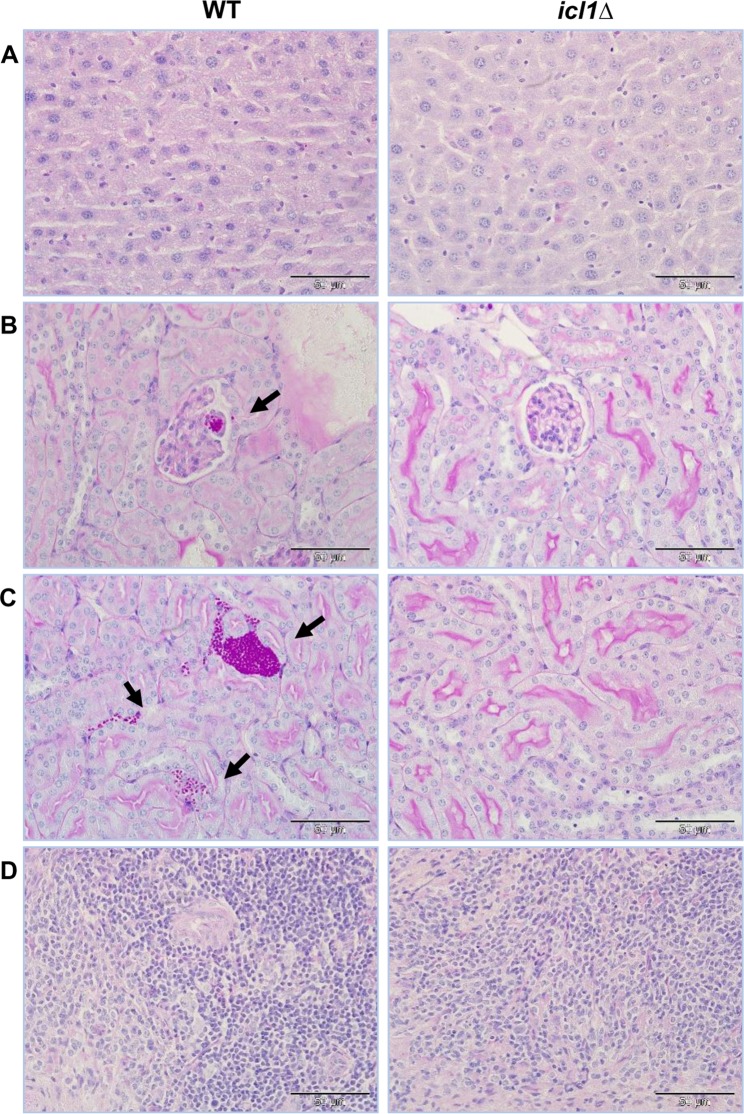
Measurements of fungal burden were performed for the C. glabrata HTL and icl1∆ strains from recovered organs at day 3 post-infection. There were no significant differences between the WT and icl1∆ strains regarding the fungal burdens in the liver (106 CFU/g tissue) and spleen (107 CFU/g tissue). However, the fungal burden in the kidney was greatly reduced in mice infected with the C. glabrata icl1∆ mutant, compared to the WT control (p < 0.05) (Fig. 5B). Indeed, the kidney fungal burden for the C. glabrata icl1∆ mutant was approximately 2 × 104 CFU/g tissue, a significant 63-fold reduction compared to the WT strain. Histopathological sections of kidneys from infected mice showed the presence of C. glabrata WT cells in glomeruli on day 3 post infection (Fig. 6). Furthermore, it appeared that C. glabrata successfully passed through glomeruli and invaded the renal cortex, as invasion of C. glabrata WT cells was observed surrounding renal tubules. In concordance with the results from the survival assay, the disruption of ICL1 rendered C. glabrata less able to invade kidney tissues in these immunosuppressed ICR mice. The MPO content in kidney homogenates was also significantly higher for mice infected with the C. glabrata WT strain (1.19 ± 0.035 µg g−1) compared to those infected with the C. glabrata icl1∆ mutant (0.93 ± 0.056 µg g−1) (Fig. 5C).
Figure 6.
Representative PAS-stained histological sections of different organs from ICR mice. Immunosuppressed ICR mice were infected with 2 × 108 C. glabrata WT and icl1∆ mutant via lateral tail vein injection, and organs were harvested on day 3 post infection (x400 magnification). (A) Liver, (B) Renal glomerulus, (C) Renal cortex, and (D) Spleen. Arrow indicate fungal cells.
Discussion
Nutrient assimilation is essential for the survival and growth of all microorganisms. Hence, for fungal pathogens to thrive in humans, they must adapt effectively to host microenvironments that offer varying nutrients availabilities. Accordingly, fungal pathogens generally display an impressive degree of metabolic flexibility, which contributes to their fitness and pathogenicity in vivo. This metabolic flexibility presents a potential target for antifungal drug discovery29. In this study, we demonstrate that the glyoxylate cycle, and in particular the enzyme isocitrate lyase, is essential for the metabolic flexibility and pathogenicity of the major pathogen C. glabrata. Taking cues from the baker’s yeast S. cerevisiae, we anticipated that ICL1 might be required in C. glabrata for the utilisation of fatty acids, ethanol and acetate. Ethanol is believed to enter the cells through passive diffusion, whereas acetate is transported to the cells through the carboxylate transporter, acetate permease30–32. Both carbon sources are converted to acetyl-CoA by acetyl-CoA synthetase. Unlike ethanol and acetate, fatty acids like oleic acid are broken down to acetyl-CoA via β-oxidation, which includes the enzymes fatty acyl-CoA oxidase, 3-hydroxyacyl-CoA dehydrogenase and 3-oxoacyl-CoA thiolase14. Acetyl-CoA fuels the glyoxylate cycle and gluconeogenesis for glucose production when glucose availability is scarce14,33.
We show that disruption of ICL1 renders C. glabrata unable to grow on acetate, ethanol and oleic acid (Figs 1 and 2). These observations were in concordance with the carbon utilisation patterns of S. cerevisiae, but they contrast with other Candida species like C. albicans28. Compared to C. glabrata, the deletion of ICL1 in C. albicans causes a more significant defect as C. albicans icl1∆ cells are unable to grow on glycerol as well28. Since C. glabrata shares a relatively recent common ancestor with many Saccharomyces species34, C. glabrata might utilise alternative carbon sources in a similar manner to S. cerevisiae rather than Candida species from the CUG clade. C. albicans lies in this CTG clade and this species requires ICL1 for the utilisation of citrate and glycerol28, in addition to fatty acids, acetate and ethanol. However, ICL deletion has no effect on the formation of biofilms by C. albicans on glucose35. Interestingly, in this study, we show that although disruption of ICL1 has no effects on the biofilm formation of C. glabrata on glucose and glycerol, it does significantly impact the biofilm formation of this fungus on acetate, lactate, ethanol and oleic acid (Fig. 3). As demonstrated by the XTT assay, these C. glabrata icl1∆ cells show reduced metabolic activity when subjected to certain alternative carbon sources. Therefore, reduced biofilm formations observed in acetate, lactate, ethanol and oleic acid-grown C. glabrata icl1∆ cells are probably attributed to the growth defect of C. glabrata icl1∆ strains, instead of the impairment of biofilm formation ability. Taken together, this shows that ICL1 is indispensable for the growth of C. glabrata in the presence of some alternative carbon sources.
Macrophages represent a first line of defence during microbial invasion and are responsible for the engulfment and killing of invading pathogens36. In this study, we show that the glyoxylate cycle is also crucial to sustain the viability of C. glabrata cells trapped within macrophages (Fig. 4). Presumably, ICL1 disruption prevents C. glabrata from reassimilating alternative carbon sources that are generated by fungal autophagy within macrophages, thereby rendering C. glabrata icl1∆ cells more susceptible to macrophage killing. This suggests that ICL1 might be an important virulence factor for C. glabrata that is required for the prolonged survival of C. glabrata cells following macrophage engulfment in vivo. Ramirez and Lorenz (2007) and others27,28 have shown that ICL1 is required for the full virulence of C. albicans in vivo. As C. glabrata is normally highly resistance to macrophage killing, we postulated that ICL1 is also crucial for alternative carbon utilisation and survival of C. glabrata in the host.
To test this, we investigated the virulence of C. glabrata icl1∆ cells using a mouse model of invasive candidiasis. ICL1 disruption significantly reduced the mortality of infected mice (Fig. 5). Furthermore, fungal burdens in the kidney were significantly reduced, while there was no significant effect of ICL1 deletion on the fungal burden load in the liver and spleen (Fig. 5). This implies that loss of ICL1 render C. glabrata less competent in kidney invasion. To establish renal candidiasis, C. glabrata transits from the renal artery via the afferent arterioles to the glomerulus in renal corpuscle and subsequently infects the renal tubules37. Histologic examination of the kidneys of infected mice further confirmed the importance of ICL1 in kidney invasion of C. glabrata (Fig. 6). The reduced virulence of C. glabrata icl1∆ cells was further supported by the reduced polymorphonuclear neutrophils (PMN) marker enzyme MPO in the kidney of infected mice (Fig. 5). Taken together, the data indicate that deletion of ICL1 leads to severe attenuation of C. glabrata virulence in mouse model of invasive candidiasis.
In conclusion, our data suggest an essential role for ICL1 in the utilisation of alternative carbon sources by C. glabrata. In addition, we suggest an important role for ICL1 in promoting the growth and prolonged survival of C. glabrata following macrophage engulfment. Most importantly, ICL1 is required for the full virulence of C. glabrata in vivo. Our results could pave a way for the development of new candidate treatments that target Icl1 for antifungal drug development. Further investigation of key metabolic enzymes and regulators of alternative carbon metabolic pathways, such as β-oxidation, glyoxylate cycle and gluconeogenesis in C. glabrata are warranted.
Materials and Methods
Strains and growth conditions
All C. glabrata strains used in this study are listed in Table 1. The triple-auxotrophic strain C. glabrata HTL (wild type, WT) was constructed from the reference strain C. glabrata ATCC 2001 through the removal of coding sequences of HIS3, LEU2 and TRP1 using a recyclable, dominant nourseothricin resistance marker SAT138. For construction of three independent strains of C. glabrata icl1∆, fusion PCR technique was used to generate gene deletion cassette containing approximately 500 bp of homologous flanking regions for ICL1, combined with nourseothricin marker gene, NAT1, barcodes identifiers and constant overlap sequences as detailed previously39,40. The C. glabrata HTL strain was transformed with ICL1 gene deletion cassette using a modified electroporation method. Nourseothricin-resistant transformants were confirmed for correct deletion of ICL1 deletion by PCR. Three independently constructed C. glabrata icl1∆ were used in this study (Table 1).
Table 1.
Candida glabrata strains used in this study.
| C. glabrata strains | Genotype | Reference |
|---|---|---|
| ATCC 2001 | Reference strain | American Type Culture Collection (ATCC) |
| HTL | Derived from ATCC 2001 his::FRT, leu2::FRT, trp1::FRT |
Jacobsen et al., 2010 |
|
icl1∆_a icl1∆_b icl1∆_c |
Derived from HTL icl1::NAT1 |
Schwarzmüller et al., 2014 |
Standard culture media were used, including YPD (Becton, Dickinson and Company, USA): yeast extract (1%, w/v), peptone (2%, w/v), glucose (2%, w/v), agar (1.5%, w/v) and YNB without amino acids (Becton, Dickinson and Company, USA): yeast nitrogen base (0.67%, w/v), ammonium sulfate (0.5%, w/v). Synthetic complete (SC) media were prepared with YNB without amino acids, supplemented with complete supplement mixture (0.2%, w/v) (Formedium, UK), glucose (2%, w/v) and agar (2%, w/v). In growth phenotype assays, glucose was replaced with other alternative carbon sources.
Growth phenotypes
Growth phenotypes of C. glabrata ATCC 2001, WT and icl1∆ strains in glucose and alternative carbon sources were investigated on SC media containing glucose (2%, w/v), sodium acetate (2%, w/v), sodium lactate (2%, v/v), ethanol (2%, v/v), glycerol (2%, v/v) or oleic acid (0.2%, w/v) (Sigma-Aldrich, USA) as the sole carbon source. A lower concentration of carbon source was used for oleic acid (0.2%, w/v) as previously described41. SC media were incubated at 37 °C for 24 to 96 h (Ramírez & Lorenz, 2007).
For spot dilution assays, C. glabrata strains were grown in YPD for overnight at 37 °C, harvested and washed twice with phosphate buffered saline (PBS), pH 7.4 before resuspended into fresh SC media (OD600nm of 1.0) with glucose, acetate, lactate, ethanol, glycerol and oleic acid. Subsequently, cell suspensions were transferred into a sterile 96-well plate and serially diluted five-fold. These dilutions were spotted on SC media supplemented with different carbon sources and incubated at 37 °C for 24 to 96 h.
For microplate-based growth assay, C. glabrata strains were grown in YPD for overnight at 37 °C, harvested and washed twice with PBS, pH 7.4 before resuspended into fresh SC media (OD600nm of 0.1) with glucose, acetate, lactate, ethanol, glycerol and oleic acid as sole carbon source. A volume of 200 µl of cell suspension was transferred into a sterile 96-well plate. Growth of C. glabrata strains was monitored for 96 h by measuring OD600nm with microtiter plate reader (Dynex Technologies, USA).
Biofilm formation
Biofilm formation of C. glabrata WT and icl1∆ mutant in different alternative carbon sources were assessed by using a modified procedure previously described42. Briefly, overnight cultures of C. glabrata WT and icl1∆ mutants were harvested and washed twice with PBS, pH 7.4 before resuspended into fresh SC media (OD600nm of 0.1) with glucose, acetate, lactate, ethanol, glycerol and oleic acid as sole carbon source. A volume of 100 µl cell suspension was dispensed into a pre-sterilized, clear and flat bottomed 96-well polystyrene cell culture plate with low-evaporation lids (Becton, Dickinson and Company, USA). The 96-well plate was covered with its original lid, sealed with parafilm and incubated for 48 h at 37 °C for biofilm formation.
The 96-well plate was washed twice with PBS, pH 7.4 and residual PBS was removed with blotting paper. Biofilm formation of C. glabrata strains was quantified by 2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) reduction assay. A volume of 100 µL solution mixture of 0.5 g/L XTT (Sigma-Aldrich, USA) and 10 mM menadione (10000: 1, v/v) (Sigma-Aldrich, USA) was added to the biofilms. The plate was covered in aluminium foil and incubated in the dark at 37 °C for 3 h. Subsequently, 80 µl of the solution was transferred to a new 96-well plate and OD490nm was measured by using a microtiter plate reader.
Fungal killing by macrophages
RAW264.7 murine macrophages were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Thermo Fisher Scientific, USA) supplemented with 10% (v/v) foetal bovine serum (FBS; Thermo Fisher Scientific, USA) and 1% (v/v) penicillin-streptomycin antibiotics (Nacalai Tesque, Japan) in cell culture flasks (Nunc; Thermo Fisher Scientific, USA) at 37 °C and 5% (v/v) CO2. The cells were seeded at a density of 5 × 105 in 12-well tissue culture plates (Becton, Dickinson and Company, USA) for 24 h at 37 °C and 5% (v/v) CO2. The cell number was determined by cell counting using a haemocytometer.
For the preparation of C. glabrata cells, overnight cultures of C. glabrata WT and icl1∆ mutant were washed and regrown to mid-exponential phase (OD600nm of 0.5) in fresh YPD. Candida glabrata cells were harvested by centrifugation, resuspended in DMEM supplemented with 10% FBS and added to RAW264.7 macrophage at a multiplicity of infection (MOI) of 1: 1 (RAW264.7: Candida). Non-phagocytosed C. glabrata cells were removed by washing with DMEM after 2 h of co-incubation. Lysates of infected RAW264.7 macrophages were harvested after 2 and 24 h of co-incubation. The cells were lysed with ice-cold sterile deionized water and plated on YPD. Colony-forming-unit (CFU) of intracellular C. glabrata cells were counted after incubation at 37 °C for 24 h. Survival ratio of phagocytosed C. glabrata cells is defined as (CFU of 24 h Sample/CFU of 2 h Control) × 100%13.
Mouse model of invasive candidiasis
The virulence of C. glabrata icl1∆ mutant in vivo was assessed using a modified murine model as previously described43. Briefly, female outbred ICR mice (6–8 weeks old, 18–20 g) were obtained from Animal Resource Unit, Faculty of Veterinary Medicine, Universiti Putra Malaysia. The mice were housed in groups of five in individually ventilated cages and offered with standard mouse cubes (Specialty Feeds, Australia) and water ad libitum. The mice were first acclimatized under controlled conditions (12/12-h light/dark cycle, 25 °C) for one week before commencement of the studies.
For survival assay, groups of 10 mice were immunosuppressed with cyclophosphamide (200 mg/kg; Merck, Germany) through intraperitoneal injection on day -3 and every fourth day thereafter. Mice were challenged intravenously via lateral tail vein on day 0 with 2 × 108 C. glabrata cells in 200 µl of saline 0.9% (w/v). Infected mice were subsequently monitored for sign of infection and humanely euthanized by cervical dislocation under anaesthesia when predetermined end-points were reached (20% body weight loss, laboured breathing, unconscious or moribund state). Survival assay was terminated at day 21 post-infection.
Fungal burdens in tissues were assayed. Groups of 5 mice were immunosuppressed with cyclophosphamide on day -3 and challenged with 2 × 108 C. glabrata cells in 200 µl of saline 0.9% (w/v). Infected mice were humanely euthanized at day 3 and organs (liver, spleen and kidney) of each mouse were procured aseptically. The organs were immediately placed in sterile, ice-cold PBS and mechanically homogenized. Subsequently, the serially diluted tissue homogenates were plated on YPD agar. CFU counts were performed after 24 h of incubation at 37 °C. All procedures involving mice were performed in accordance to the protocols approved by the Institutional Animal Care and Use Committee (IACUC), Universiti Putra Malaysia (ethical approval number: UPM/|ACUC/AUPR-034/2017).
Histology
Harvested organs from infected mice were fixed and kept and in 10% neutral buffered formalin until processed for histology. Fixed organs were paraffin-embedded, sectioned at 5 µm, and stained with periodic acid-Schiff (PAS) according to standard staining protocols. Histological samples were viewed and analysed with an Olympus BX51TRF microscope (Olympus Corporation, Japan).
Myeloperoxidase quantification
Kidney homogenates of the infected mice were centrifuged twice for 5 min at 4 °C (5000 × g) and the supernatants were stored at −80 °C until myeloperoxidase (MPO) quantification. MPO contents were determined by the commercially available mouse MPO enzyme-linked immunosorbent assay (ELISA) kit (Fine Biotech Co., China) according to the manufacturer’s recommendations.
Statistical analyses
Statistical analyses were performed using GraphPad Prism Version 7.0 Software (GraphPad Software Inc., USA). All experiments were performed at least in three replicates and all data were expressed as mean values from all replicates with the corresponding standard deviations (SD). Differences between control (WT) and sample (mutant) were assessed by unpaired t-test and a p < 0.05 was considered to be statistically significant. All significant differences were indicated in the figures, with *, **, and ***indicating p < 0.05, <0.01 and <0.001. Comparison and statistical analysis of survival curves was performed using Mantel-Cox log rank test.
Acknowledgements
We would like to acknowledge Professor Karl Kuchler from Medical University of Vienna for the kind gifts of C. glabrata strains used in this study. This study was funded by Fundamental Research Grant Scheme (FRGS) from Ministry of Education (MOE), Malaysia (Grant number: 01-01-14-1456FR). S.Y. is a recipient of the MyBrain 15 Scholarship from MOE, Malaysia. A.B. was supported by the Medical Research Council Centre for Medical Mycology at the University of Aberdeen (MR/N006364/1), by a programme grant from the UK Medical Research Council (MR/M026663/1), by a Strategic Award from the Wellcome Trust (097377) and by a grant from the UK Biotechnology and Biological Sciences Research Council (BB/P020119/1).
Author Contributions
S.Y., L.T., A.B., K.L., Y.K. and D.S. designed the experiments. S.Y. executed the experiments with minor assistance by T.S. S.Y. analysed and interpreted the data. S.Y. and L.T. wrote the manuscript with inputs from A.B., K.L. and Y.K. All authors gave final approval for publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arendrup MC. Epidemiology of invasive candidiasis. Curr Opin Crit Care. 2010;16(5):445–452. doi: 10.1097/MCC.0b013e32833e84d2. [DOI] [PubMed] [Google Scholar]
- 2.Kullberg BJ, Arendrup MC. Invasive candidiasis. N Engl J Med. 2015;373(15):1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 3.Sobel JD. The emergence of non-albicans Candida species as causes of invasive candidiasis and candidemia. Curr Infect Dis Rep. 2016;8:427–433. doi: 10.1007/s11908-006-0016-6. [DOI] [PubMed] [Google Scholar]
- 4.Beardmore RE, et al. Drug-mediated metabolic tipping between antibiotic resistant states in a mixed-species community. Nat Ecol Evol. 2018;2(8):1312–1320. doi: 10.1038/s41559-018-0582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018;73:i4–i13. doi: 10.1093/jac/dkx444. [DOI] [PubMed] [Google Scholar]
- 6.Wächtler B, et al. Candida albicans-epithelial interactions: Dissecting the roles of active penetration, induced endocytosis and host factors on the infection process. PLOS One. 2012;7(5):e36952. doi: 10.1371/journal.pone.0036952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moyes DL, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532(7597):64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunke S, Hube B. Two unlike cousins: Candida albicans and C. glabrata infection strategies. Cell Microbiol. 2013;15(5):701–708. doi: 10.1111/cmi.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duggan S, et al. Neutrophil activation by Candida glabrata but not Candida albicans promotes fungal uptake by monocytes. Cell Microbiol. 2015;17(9):1259–1276. doi: 10.1111/cmi.12443. [DOI] [PubMed] [Google Scholar]
- 10.Seider K, et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J Immunol. 2011;187(6):3072–3086. doi: 10.4049/jimmunol.1003730. [DOI] [PubMed] [Google Scholar]
- 11.Seider K, et al. Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot Cell. 2014;13(1):170–183. doi: 10.1128/EC.00262-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roetzer A, Gratz N, Kovarik P, Schüller C. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol. 2010;12(2):199–216. doi: 10.1111/j.1462-5822.2009.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng TS, et al. SNF3 as high affinity glucose sensor and its function in supporting the viability of Candida glabrata under glucose-limited environment. Front Microbiol. 2015;6:1334. doi: 10.3389/fmicb.2015.01334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3(5):1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur R, Ma B, Cormack BP. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci USA. 2007;104(18):7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueno K, et al. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLOS One. 2011;6(9):e24759. doi: 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunha DV, Salazar SB, Lopes MM, Mira NP. Mechanistic insights underlying tolerance to acetic acid stress in vaginal Candida glabrata clinical isolates. Front Microbiol. 2017;28(8):259. doi: 10.3389/fmicb.2017.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lorenz MC, Fink GR. Life and death in a macrophage: Role of the glyoxylate cycle in virulence. Eukaryot Cell. 2002;1(5):657–662. doi: 10.1128/EC.1.5.657-662.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology. 2009;155(10):3166–3175. doi: 10.1099/mic.0.030858-0. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz-Elías EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11(6):638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang FC, Libby SJ, Castor ME, Fung AM. Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect Immun. 2005;73(4):2547–2549. doi: 10.1128/IAI.73.4.2547-2549.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall DM, Duffy PS, Dupont C, Prescott JF, Meijer WG. Isocitrate lyase activity is required for virulence of the intracellular pathogen Rhodococcus equi. Infect Immun. 2005;73(10):6736–6741. doi: 10.1128/IAI.73.10.6736-6741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindsey TL, Hagins JM, Sokol PA, Silo-Suh LA. Virulence determinants from a cystic fibrosis isolate of Pseudomonas aeruginosa include isocitrate lyase. Microbiology. 2008;154(6):1616–1627. doi: 10.1099/mic.0.2007/014506-0. [DOI] [PubMed] [Google Scholar]
- 24.Ebel F, et al. Analysis of the regulation, expression, and localisation of the isocitrate lyase from Aspergillus fumigatus, a potential target for antifungal drug development. Fungal Genet Biol. 2006;43(7):476–489. doi: 10.1016/j.fgb.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Schobel F, et al. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect Immun. 2007;75(3):1237–1244. doi: 10.1128/IAI.01416-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rude TH, Toffaletti DL, Cox GM, Perfect JR. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun. 2002;70(10):5684–5694. doi: 10.1128/IAI.70.10.5684-5694.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barelle CJ, et al. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8(6):961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez MA, Lorenz MC. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot Cell. 2007;6(2):280–290. doi: 10.1128/EC.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ene IV, Brunke S, Brown AJ, Hube B. Metabolism in fungal pathogenesis. Cold Spring Harb Perspect Med. 2014;4(12):a019695. doi: 10.1101/cshperspect.a019695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paiva S, Devaux F, Barbosa S, Jacq C, Casal M. Ady2p is essential for the acetate permease activity in the yeast Saccharomyces cerevisiae. Yeast. 2004;21(3):201–210. doi: 10.1002/yea.1056. [DOI] [PubMed] [Google Scholar]
- 31.Casal M, Paiva S, Queirós O, Soares-Silva I. Transport of carboxylic acids in yeasts. FEMS Microbiol Rev. 2008;32(6):974–994. doi: 10.1111/j.1574-6976.2008.00128.x. [DOI] [PubMed] [Google Scholar]
- 32.Vieira N, et al. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol Microbiol. 2010;75(6):1337–1354. doi: 10.1111/j.1365-2958.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turcotte B, Liang XB, Robert F, Soontorngun N. Transcriptional regulation of nonfermentable carbon utilization in budding yeast. FEMS Yeast Res. 2010;10(1):2–13. doi: 10.1111/j.1567-1364.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dujon B, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 35.Ishola OA, et al. The role of isocitrate lyase (ICL1) in the metabolic adaptation of Candida albicans biofilms. Jundishapur J Microbiol. 2016;9(9):e38031. doi: 10.5812/jjm.38031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert AS, Wheeler RT, May RC. Fungal pathogens: survival and replication within macrophages. Cold Spring Harb Perspect Med. 2015;5(7):a019661. doi: 10.1101/cshperspect.a019661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher JF, Kavanagh K, Sobel JD, Kauffman CA, Newman CA. Candida urinary tract infection: pathogenesis. Clin Infect Dis. 2011;52(6):437–451. doi: 10.1093/cid/cir110. [DOI] [PubMed] [Google Scholar]
- 38.Reuß O, Vik Å, Kolter R, Morschhäuser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 39.Noble SM, Johnson AD. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell. 2005;4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarzmüller T, et al. Systematic phenotyping of a large-scale Candida glabrata deletion collection reveals novel antifungal tolerance genes. PLOS Pathog. 2014;10(6):e1004211. doi: 10.1371/journal.ppat.1004211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piekarska K, et al. Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell. 2006;5(11):1847–1856. doi: 10.1128/EC.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pierce CG, et al. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3(9):1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calcagno AM, et al. Candida glabrata STE12 is required for wild-type levels of virulence and nitrogen starvation induced filamentation. Mol Microbiol. 2003;50(4):1309–1318. doi: 10.1046/j.1365-2958.2003.03755.x. [DOI] [PubMed] [Google Scholar]