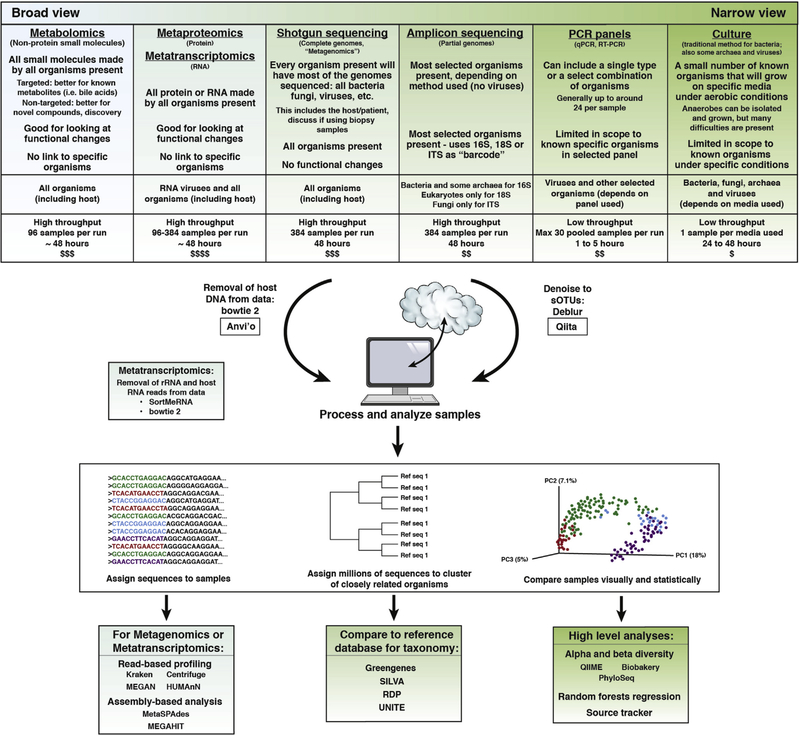
Figure 4.
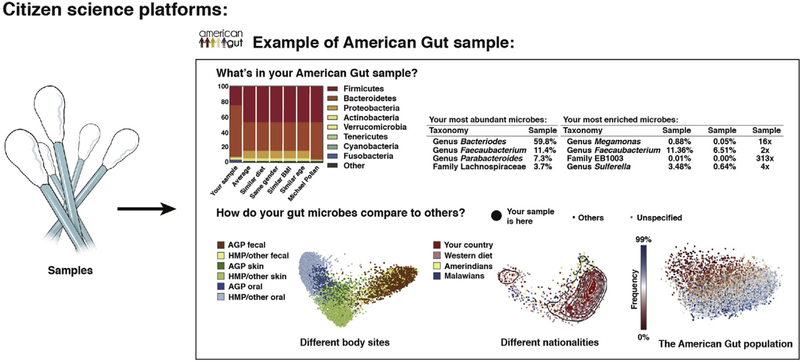
Once samples are collected, the samples can be put through molecular preparations and DNA sequencing to generate microbiome data. Two common types of protocols are amplicon sequencing and shotgun sequencing. In amplicon sequencing, PCR primers are used to target a specific region of a specific gene, focusing sequencing effort on just those fragments. One of the most widely used protocols targets the V4 region of the 16S rRNA gene.24 In shotgun sequencing, the DNA in the sample is randomly sheared and sequenced, generating data from many different parts of the genome. The specifics of the molecular protocol used before shotgun sequencing are important for what type of data are being examined, and this type of sequencing can be used, for example, for metagenomics and metatranscriptomics. The initial processing performed on the data after sequencing depends on the type of sequencing performed. For amplicon studies, one common strategy is to upload the data into Qiita81 and to use Deblur82 to resolve sequence data into single-sequence variants called suboperational taxonomic units (sOTUs). Taxonomic assignments generally are performed using naive Bayes classifiers such as the RDP classifier,59 as implemented in the q2-feature-classifier against reference databases such as Greengenes,83 SILVA,78 RDP,79 or UNITE84 (fungal internal transcribed spacer [ITS]) depending on the amplicon target. Shotgun sequencing of host-associated samples first requires preprocessing to remove either host DNA before analysis. Typically, the shotgun data then are summarized using tools such as Kraken,75 MEGAN,85 or HUMAnN286 to generate taxonomic or functional profiles, or are assembled with tools such as metaSPAdes87 and MEGAHIT.88 For both sequencing methods, higher-level analyses (eg, α and β diversity, taxonomic profiling, and machine learning) subsequently are used to assay patterns of microbiome variation in the context of the study design. Metagenomic assemblies also can be analyzed through platforms such as Anvi’o.89 SourceTracker,90 a Bayesian estimator of the sources that make up each unknown community, is useful for classifying microbial samples according to the environment of origin.91 Citizen Science platforms, such as the American Gut Project,25 standardize the molecular work and bioinformatic processing to generate a basic summary report of the content of an individuals sample. In the case of the American Gut Project, the samples also are placed into the context of a few other popular microbiome studies through data integration.