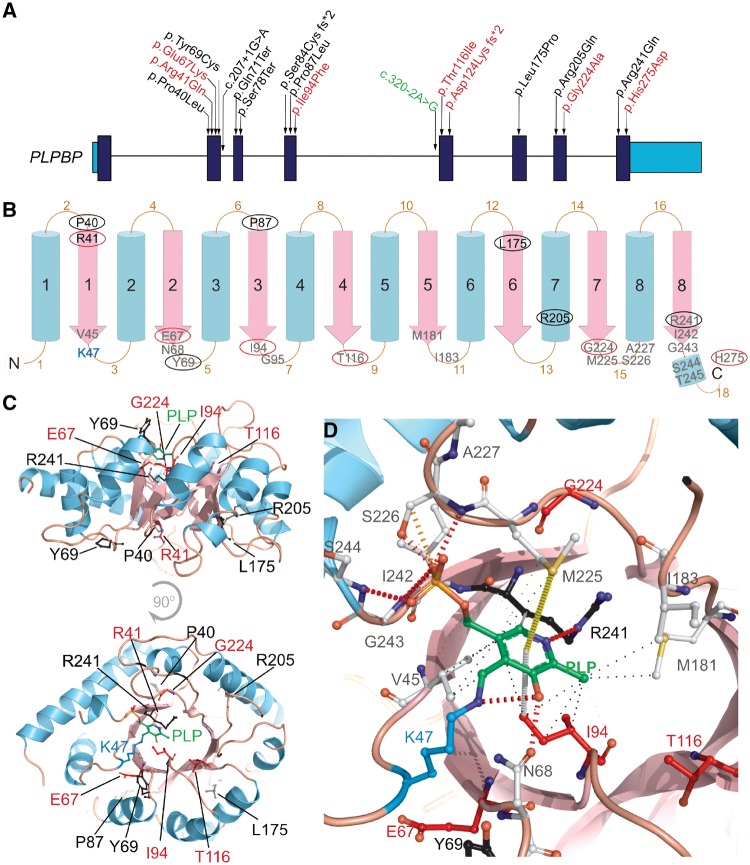
Figure 2.
Pathogenic variants reported so far and their genetic location, predicted secondary structure and 3D structure in the PLPHP protein in the context of PLP-binding. (A) Human PLPBP gene structure, protein coding exons shown in dark blue and 5′ and 3′ UTR shown in light blue. Position of the variants reported previously by Darin et al. (2016) and Plecko et al. (2017) are shown in black, seven novel variants identified by this study are shown in red and a splicing variant reported previously but also observed in our cohort is shown in green. (B) 2D graphical representation of the PLPHP protein based on secondary structure prediction and the tridimensional model (shown in D). Blue cylinders represent the outer α-helices and pink arrows represent the inner β-strands that comprise the (β/α)8-TIM barrel structure. Residues observed mutated in PLPHP-deficiency are shown in circles, black for variants reported previously or red for novel variants reported here. Residues located within 6Å of the modelled PLP position are shown in grey. (C) 3D structure of the human PLPHP model showing the PLP molecule in green, the lysine 47 residue in blue and the positions of the residues found mutated in PLPHP deficiency in black or red according to A. (D) Predicted PLP-binding pocket showing the key lysine 47 (K47) as a PLP-Lys adduct (blue and green), PLP-pocket residues (<6Å radius) and the residues found mutated in PLPHP deficiency in black or red according to A. Non-covalent contacts as calculated by Arpeggio are shown; black dashed lines indicate hydrophobic interactions, orange and red dashed lines represent weak and strong hydrogen bonds, grey dashed line represents carbon-pi interaction and a yellow dashed line indicates a methyl-sulphur-pi interaction. Note that the variant p.His275Asp was co-inherited homozygously with p.Thr116Ile in Patient 1, we report this as a variant of unknown significance.