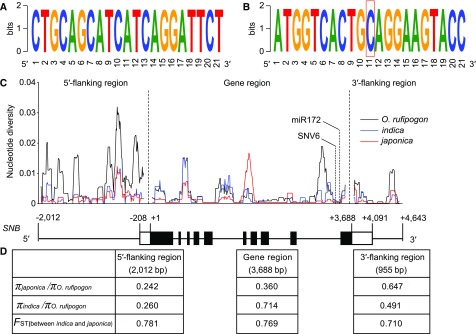
Figure 10.
Nucleotide Diversity and indica-japonica Differentiation Analysis of SNB using 46 Accessions of O. rufipogon, 48 indica Varieties, and 38 japonica Varieties.
(A) Conservation analysis showing that both wild and cultivated rice lines have identical sequences at the miR172 target site.
(B) Conservation analysis of the 10-bp upstream and downstream sequences surrounding the SNP SNV6, showing that both wild and cultivated rice have a conserved C at the SNV6 site. The SNP SNV6 is boxed in red.
(C) Sliding-window analysis of nucleotide polymorphism (π) in SNB. The values were calculated for each sliding window of 100 bp with an increment of 10 bp. Black boxes represent exons, white boxes represent UTRs, and thin black lines indicate flanking regions and introns.
(D) The values of πjaponica/πO. rufipogon, πindica/πO. rufipogon, and FST (between indica and japonica) were each calculated for the 5′-flanking region, gene region, and 3′-flanking region of SNB.