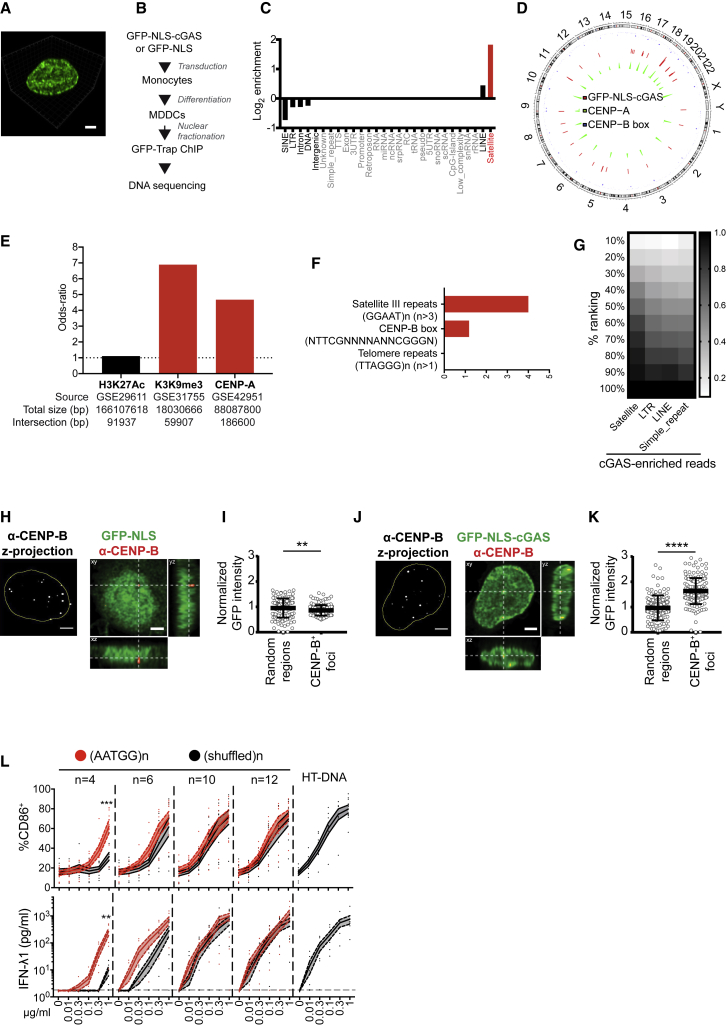
Figure 5.
Nuclear cGAS Associated with Centromeric Satellite DNA
(A) 3D projection of the nucleus of a DC expressing GFP-NLS-cGAS (green).
(B) Experimental scheme for ChIP-seq of GFP-NLS-cGAS stably transduced in DCs.
(C) Annotation of the filtered peaks of the ChIP-seq on GFP-NLS-cGAS. Elements with <10 peaks are grayed out (1 donor representative of 3 independent donors).
(D) Circular plot showing the distribution of GFP-NLS-cGAS and of CENP-A peaks and localization of CENP-B box (consensus sequence) on the hg38 genome. The cGAS track represents the fold change (chip over input) of selected filtered peaks (162 regions) from the 3 donors. The CENP-A track represents the density of CENP-A intersection peaks (5,977 regions) computed on windows of size 107 across the genome. The CENP-B box track reports on the x axis the genomic position of the region (occurrence of CENP-B box consensus sequence) and on the y axis the minimal distance (log10 transformed) of the region to its two neighboring regions.
(E) Association of GFP-NLS-cGAS peaks with public H3K27Ac peaks from GM12878 cells, H3K9me3 peaks from peripheral blood mononuclear cells (PBMCs), and endogenous CENP-A peaks from HeLa S3 cells. Filtered GFP-NLS-cGAS peaks for donor 1 are used (404 peaks, 1,545,600 bp) (representative of 3 independent donors).
(F) Sequence enrichment in cGAS-specific peaks from GFP-NLS-cGAS ChIP-seq over GFP-NLS ChIP-seq filtered peaks (intersection of peaks from two independent donors). Three motifs were assessed: satellite III DNA motif repeats, [GGAAT]n > 3; CENP-B box consensus sequence, NTTCGNNNNANNCGGGN; and telomeric repeats, [TTAGGG]n > 1.
(G) cGAS-specific read enrichment on repeats. A repeat occurrence is considered if the read count per million (cpm) is ≥2 in any sample. Repeats are grouped into n = 10 bins according to the ChIP read enrichment over GFP-NLS. For each repeat class R, the fraction of occurrences within the first i bins, corresponding to the top (100/i)% ranks, is shown as a gradient from white to black. Only repeat classes with at least 10 read occurrences in the genome and that pass the cpm cutoff are considered and sorted from left to right by decreasing number of occurrences in the top 50% (1 donor representative of n = 2 independent donors).
(H) DC stably transduced with GFP-NLS lentivector in pTRIP-SFFV and stained for CENP-B. (Left) Z-projection of CENP-B (white) with nuclear mask (yellow) and (right) orthogonal projections (single confocal plane) of CENP-B (red) and GFP-NLS (green). Scale bar, 2 μm.
(I) Quantification of GFP intensity in CENP-B foci or random regions in the nucleus, normalized over mean nuclear GFP intensity, in cells transduced as in (H); n ≥ 140 foci or random regions in 7 independent cells. Each dot represents a single CENP-B focus. Means and SDs are represented. One donor representative of n = 4 donors in 2 independent experiments. Student’s t test.
(J) DC stably transduced with GFP-NLS-cGAS lentivector in pTRIP-SFFV and stained for CENP-B, shown as in (H). Scale bar, 2 μm.
(K) Quantification of GFP-NLS-cGAS intensity in CENP-B foci or random regions in the nucleus as in (I).
(L) CD86 and IFN-λ1 expression by DCs transfected with synthetic DNA repeats coding for the AATGG satellite motif, the corresponding shuffled sequence, or HT-DNA at the indicated DNA concentrations (solid lines, means; dotted lines, SEMs; independent donors: n = 9 for CD86, n = 7 for IFN-λ1; 2-way ANOVA with Tukey test on log-transformed data for IFN-λ1).
∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. See also Figure S4.