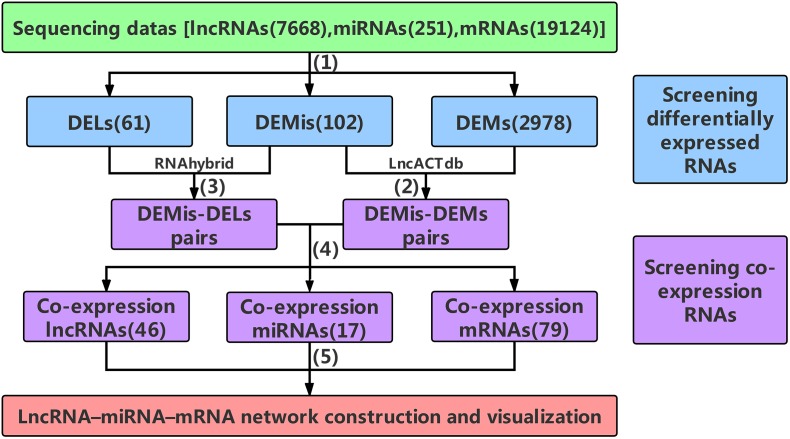
FIGURE 1.
The main frame of data processing. (1) High-throughput sequencing data were pre-selected according to the | log2FC| ≥ 4(| log2FC| ≥ 2 for DEMs) and P-values < 0.001. (2) The DEMis-DEMs pairs were obtained using LncACTdb. (3) According to MFE theory, the DEMis-DELs pairs were screened using the RNAhybrid program. (4) the DEMis-DEMs pairs were merged with the DEMis-DELs pairs to obtain the co-expression RNAs. (5) 46 co-expression lncRNAs, 17 co-expression miRNAs, and 79 co-expression mRNAs were finally selected to construct the lncRNA–miRNA–mRNA network. DELs, differentially expressed lncRNAs; DEMis, differentially expressed miRNAs; DEMs, differentially expressed mRNAs; MFE, minimum free energy.