FIG 4.
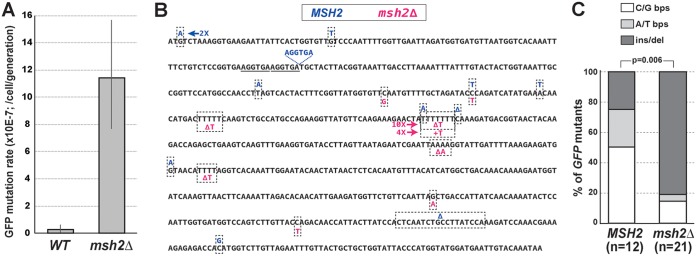
Fluorescence-based mutation reporter captures the mutator phenotype and mutational signature of the msh2Δ DNA mismatch repair mutant in C. glabrata. (A) Using CRISPR, the yEGFP-NAT cassette was integrated into the same genetic locus in an ATCC 2001-derived strain carrying a deletion of MSH2. The mutation rate of yEGFP was measured and found to be increased 40-fold by msh2Δ relative to the strain carrying wild-type MSH2. Error bars, 95% confidence intervals. (B and C) Sequencing of yEGFP in nonfluorescent mutants isolated by FACS from MSH2 and msh2Δ strains identified a number of mutations throughout the yEGFP ORF. Whereas mutations in the MSH2 strain were mostly base pair substitutions, the majority of mutations in the msh2Δ strain were single nucleotide frameshifts (insertions or deletions) in mononucleotide runs, recapitulating the mutational signature of msh2Δ in S. cerevisiae (37, 38, 41). bps, base pair substitution; ins/del, insertion or deletion.