Abstract
Ubiquitin and ubiquitin chains are recognized by a large and growing family of receptor proteins. NMR spectroscopy provides a powerful means to evaluate whether and how a protein binds to ubiquitin. It can be used to measure binding affinities, to map interaction surfaces, and to solve the three-dimensional structure of ubiquitin:receptor complexes. Herein, we describe three NMR techniques of varying complexity that are valuable tools to characterize protein:protein complexes. These include heteronuclear correlation experiments, paramagnetic relaxation enhancement (PRE) experiments via spin labeling, and techniques designed to obtain intermolecular dipole–dipole interactions by nuclear Overhauser effects (NOEs).
Keywords: Intermolecular NOEs, NMR, Paramagnetic relaxation enhancement, Protein interactions, Ubiquitin, Ubiquitin receptor
1. Introduction
Ubiquitylation leads to new or altered interactions with proteins or complexes that contain ubiquitin-binding domains (1), which are abundant and diverse (2). Ubiquitin receptors recognize ubiquitin through helices, β-strands or loops, and thus, experimental methods are needed to map ubiquitin-binding surfaces. NMR spectroscopy is a powerful method for characterizing protein:protein interactions (3, 4). Recent advancements have enabled its use for large complexes, such as proteasome (5–7). Here, we describe three NMR-based methods to determine whether a protein is a ubiquitin receptor and if so, to define how ubiquitin recognition occurs.
2. Materials
15N labeled NH4Cl (98% 15N), D-glucose- 13C6 (99% 13C), D-glucose-1,2,3,4,5,6,6-d7 (97% 2H), deuterium oxide (99.9% 2H, 1 kg/bottle), and deuterium oxide (99.96% 2H, 0.5 mL/capsule) (Sigma-Aldrich, St. Louis, MO).
10× M9 salts (1,000 mL): Add 67.8 g Na2HPO4, 30.0 g KH2PO4, and 5.0 g NaCl to 800 mL of distilled H2O, stir until dissolved, and then adjust to 1,000 mL with distilled H2O. Sterilize by autoclaving and store at 4°C for up to 2 months.
M9 minimal media (1,000 mL): Dissolve 1.0 g NH4Cl in 100 mL of 10× M9 salts and 880 mL of distilled H2O. Sterilize by autoclaving. Prior to inoculation, add 20 mL of 20% (w/v) D-glucose (sterile), 2 mL of 1 M MgSO4 (sterile), 0.5 mL of 1% (w/v) Thiamine–HCl (sterile), 0.1 mL of 1 M CaCl2 (sterile), and appropriate antibiotic.
99.9% Deuterated M9 minimal media (1,000 mL): Dissolve 1.0 g NH4Cl, 6.78 g Na2HPO4, 3.0 g KH2PO4, 0.5 g NaCl in 980 mL of 99.9% D2O, and sterilized by filtering with a 0.22 μm PVDF membrane. Prior to inoculation, add 4 g D-glucose-1,2,3,4,5,6,6-d7 (see Note 1), 2 mL of 1 M MgSO4, 0.5 mL of 1% (w/v) Thiamine–HCl, 0.1 mL of 1 M CaCl2, and appropriate antibiotic. All additives should be dissolved in 99.9% D2O and sterilized by filtering with a 0.22 μm membrane (see Note 2).
Isopropyl β-D-thiogalactoside (IPTG): Dissolve in 99.9% D2O and sterilize by filtering with a 0.22 μm membrane when added to 99.9% deuterated M9 minimal media.
Ubiquitin: 15N or 13C isotopically enriched ubiquitin with a C-terminal polyhistidine tag can be expressed in Escherichia coli BL21(DE3) cells and purified by Ni-NTA resin followed by size exclusion chromatography on a fast protein liquid chromatography (FPLC) system. Unlabeled ubiquitin is available commercially. K48-linked or K63-linked ubiquitin chains are produced as described (8–10). Methods for producing K11-linked chains (11), linear ubiquitin chains (12), or atypical diubiquitin linkages (K6, K27, K29, and K33) (13, 14) are also available.
Isotopically labeled and unlabeled ubiquitin receptors can be produced by overexpression in E. coli and purified by affinity chromatography, most commonly using a GST or histidine tag, followed by size exclusion chromatography on an FPLC system.
Buffer 1 (ideal for NMR experiments): 20 mM sodium phosphate buffer, pH 6.5 (see Note 3), 50 mM NaCl (see Note 4), 0.1% NaN3, and 10% D2O (see Note 5). A reducing agent, such as 4 mM DTT, can be included for proteins with cysteines (ubiquitin has none).
Typical protein sample concentrations are 0.5 mM and a sample volume of 250 μL is sufficient for a conventional Shigemi tube (see Note 6).
MTSL: (1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfonate (Toronto Research Chemicals Inc., Ontario, Canada).
TEMPO-maleimide: N-(1-oxyl-2,2,6,6-tetramethyl-4-piperidinyl) maleimide (Toronto Research Chemicals Inc., Ontario, Canada).
DTNB: 5,5′-dithiobis(2-nitrobenzoic acid) (TCI America, Portland, OR).
L-Ascorbic acid.
3. Methods
In this section, we outline (Subheading 3.1) how to prepare protein labeled for NMR studies, (Subheading 3.2) the use of 2D heteronuclear correlation experiments to determine whether a protein binds ubiquitin and the binding constant for this interaction, the use of (Subheading 3.3) paramagnetic spin labeling and (Subheading 3.4) nuclear Overhauser effect spectroscopy (NOESY) experiments to study ubiquitin:receptor complexes, and (Subheading 3.5) how to use NMR data to calculate three-dimensional structures of ubiquitin:receptor complexes.
3.1. Preparing Isotopically Labeled Protein for NMR Studies
An important strategy in the use of NMR to study protein:protein interactions is the production of samples labeled with NMR active isotopes, which include 15N, 13C, and 2H. Recently, such samples have been produced with cell-free (15) or yeast expression systems, including Pichia pastoris (16) and Kluyveromyces lactis (17). The most common and affordable approach however is to use E. coli systems with an IPTG-inducible T7 RNA polymerase promoter to overexpress the protein to be labeled, as outlined below. To ease purification, the protein is expressed from an antibiotic resistant vector in frame with an affinity tag that can be removed by using an enzymatic cleavage site.
Transform E. coli and generate a starter culture. E. coli transformed with an expression vector of interest is grown overnight on LB plates containing the appropriate antibiotic for selection. Resulting colonies are shaken overnight, or to confluency, in 10 mL of LB media supplemented with appropriate antibiotic to generate a starter culture.
Grow E. coli in M9 minimal media to produce isotopically labeled sample. A 10 mL starter culture grown in LB media can be used directly to inoculate 1 L of M9 minimal media containing the appropriate antibiotic or to increase isotope labeling efficiency, the cells can be gently pelleted at 2,000×g for 20 min prior to suspension in the M9 minimal media. Fernbach flasks allow for appropriate aeration of 1 L cultures. For 15N and/or 13C labeling, 1 g/L of 15N labeled NH4Cl and/or 4 g/L of 13C labeled glucose (see Note 1) is used as the only nitrogen and/or carbon source, respectively; for 2H labeling, H2O is substituted with D2O. 2H labeling requirements range from 50 to 100%, depending on the NMR experiment. Special adaptation is typically required to produce fully deuterated samples (see Note 7). Cells grow more slowly in M9 minimal media than LB and typically take 5–7 h to reach the optimal cell density of OD600 ~ 0.5. Protein expression conditions vary depending on the protein, but often 0.2–1.0 mM IPTG is used either for 3–4 h at 37°C or overnight at a lower temperature, such as 16°C.
Purify protein for NMR. Protein samples for NMR experiments are purified with standard methods. Affinity chromatography followed by size exclusion chromatography on an FPLC system offers a convenient and effective purification scheme. Before starting NMR data collection, it may be necessary to optimize salt concentration, pH, and temperature (see Note 8) to ensure the stability of a ubiquitin receptor throughout the duration of the planned experiment. Ubiquitin is stable in a variety of buffer conditions including Buffer 1. For NMR experiments that require the removal of the H2O signal, H2O and the exchangeable hydrogens within the proteins are replaced with 2H by lyophilization. After purification, samples are flash frozen with liquid nitrogen and then subjected to lyophilization until they are in a dry powder form. Resuspension is performed with 99.96% D2O.
3.2. Two-Dimensional Heteronuclear Correlation Experiments to Study Ubiquitin Receptors
3.2.1. Test Whether a Protein Binds Ubiquitin by NMR
Heteronuclear experiments that correlate hydrogen atoms to their attached nitrogen or carbon can be used to test whether a protein binds ubiquitin, to map contact surfaces when binding does occur, and in some cases, to provide a binding constant for the interaction. When the structure of the putative ubiquitin-binding domain is known, these experiments can sometimes enable the modeling of a 3D structure of the complex. They require less protein sample compared to other NMR experiments, and the TROSY version (18) of this class of experiment enables the study of large complexes, such as the 670 kDa core particle of proteasome from Thermoplasma acidophilum (5–7).
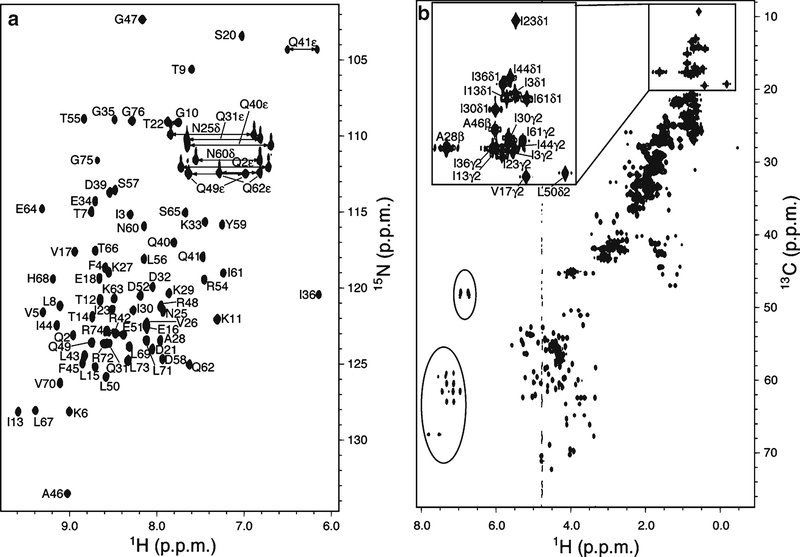
2D 1H, 15N correlation experiments display amide signals in an 15N labeled protein enabling every amino acid except proline to be readily monitored, as displayed for ubiquitin in Fig. 1a. Similarly, 2D 1H, 13C correlation experiments display all aliphatic and aromatic protons, as displayed in Fig. 1b for ubiquitin. Both versions of this experiment are easy to execute, as they are part of the software system installed on a modern NMR spectrometer. They generally take <1 h and example parameters are provided in Table 1 for 1H, 15N heteronuclear single quantum coherence (HSQC, Experiment 1) and 1H, 13C heteronuclear multiple quantum coherence (HMQC, Experiment 2) experiments acquired on ubiquitin. The most difficult aspect in the application of this technique is assigning NMR signals to specific atoms within the protein. Standard methods exist for this task (19, 20) however and ubiquitin has been well studied by NMR with its chemical shift assignments published [Biological Magnetic Resonance data Bank (BMRB) (http://www.bmrb.wisc.edu/) Entry 6457].
Fig. 1.
1H, 15N/ 13C correlation experiments for ubiquitin. (a) 1H, 15N HSQC spectrum of the distal subunit of K48-linked diubiquitin with its lysine at position 48 substituted with arginine. (b) 1H, 13C HMQC spectrum of the proximal subunit of K48-linked diubiquitin. Parameters for each spectrum are listed in Table 1. The resonances of 1H (a) or dispersed methyl groups (b) are labeled based on ubiquitin chemical shift data from BMRB Entry 6457 (http://www.bmrb.wisc.edu/). The circled resonances in (b) are from folded aromatic groups.
Table 1.
Parameters of NMR experiments for studying ubiquitin and its complexes with S5a (196–306) used as an example receptor
| Experiment | Sample | Parameters | Detection | |
|---|---|---|---|---|
| 1 15N HSQC (Fig. 1a) | 15N ubiquitin | Shared | Additional | 1H–15N signals |
| CH = 4.773 ωH = 12.5 at = 0.1 d1 = 1.0 np = 1,000 nt = 16 CH = 4.773 at = 0.1 |
CN = 118.08 ωN = 36 niN = 40 |
|||
| 2 13C HMQC (Fig. 1b) | 13C ubiquitin | CC = 42.484 ωC = 70.5 niC = 128 |
1H–13C signals | |
| 3 3D 13C-filtered, 13C-edited NOESY (Fig. 5a) | 13C S5a (196–306): unlabeled ubiquitin | ωH = 12.5a CC = 28.778 ωC = 38.8 d1 = 1.0 mix = 0.1 np = 740 nt = 16 niH = 80 niC = 64 |
Intermolecular sidechain NOEs | |
| 4 3D 15N-dispersed NOESY (Fig. 5b) | 2H, 15N S5a (196–306): unlabeled ubiquitin | ωH = 11.5 C N = 119.49 ωN = 26.6 d1 = 1.5 mix = 0.2 np = 950 nt = 8 niH = 140 niN = 54 |
Intermolecular NOEs between NH of 2H, 15N protein and all 1H of unlabeled protein; also intramolecular NH- NH NOEs | |
| 5 4D 15N-edited, 13C-dispersed NOESY (Fig. 5c) | 2H, 15N S5a (196–306):13C ubiquitin | ωH = 11.5 CC = 20.001 ωC = 40 d1 = 1.5 mix = 0.2 np = 920 nt = 16 niH = 80 niC = 64 |
Intermolecular NH-to-sidechain NOEs | |
CH, CN, and CC : carrier frequency of 1H, 15N and 13C (ppm). ωH, ωN, and ωC : sweep width of 1H, 15N and 13C (ppm). at: acquisition time (s). d1: delay time between scans (s). np: number of data points acquired in the FID. nt: number of transients. niH, niN and niC : number of increments in the indirect 1H, 15N and 13C dimension. mix: mixing time (s)
The sweep width in indirect 1H dimension can be adjusted to a smaller value
Binding events cause spectral changes, including shifting of NMR signals due to changes in chemical environment or loss of signal, due to the formation of a large complex or chemical exchange between the free and bound state. In executing 2D heteronuclear correlation experiments to test for protein:protein interactions, it is generally best to have one protein isotopically labeled and the other unlabeled to reduce spectral crowding, as outlined below. Since amino acid assignments are available for ubiquitin and it gives well-dispersed NMR signals (Fig. 1), it is convenient to label and observe its spectral changes upon addition of unlabeled receptor, as described below and demonstrated in Fig. 2a for its binding to hRpn13 (see Note 9).
Fig. 2.
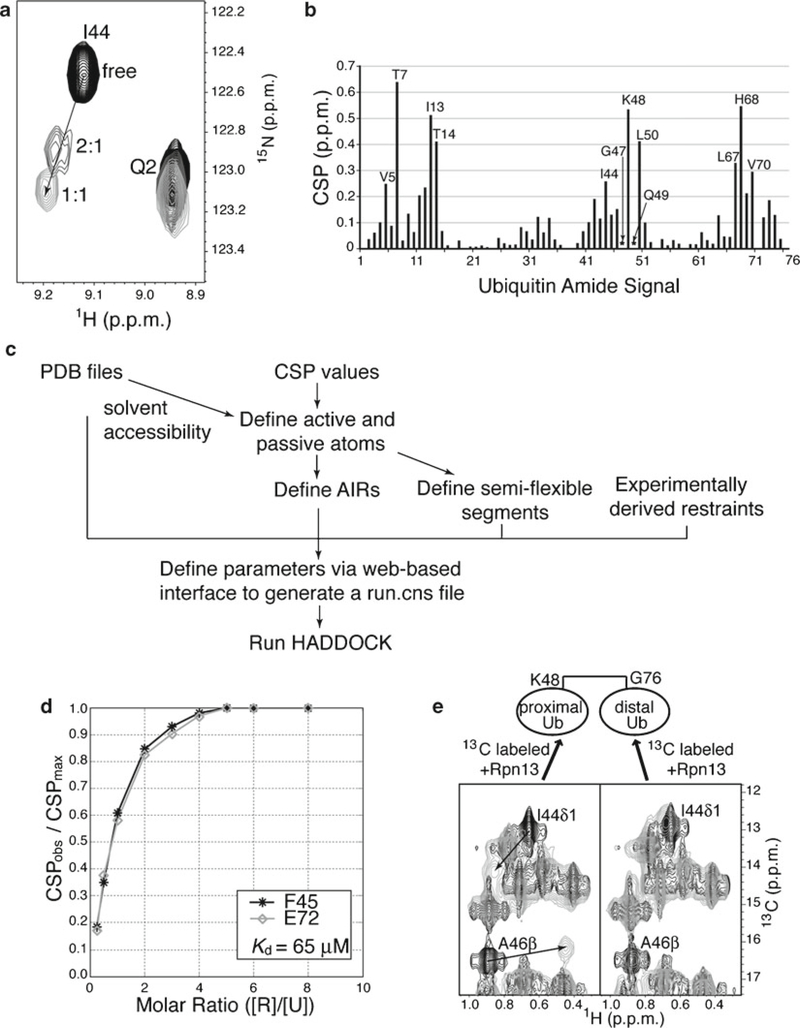
Use of chemical shift changes to detect binding between ubiquitin and a receptor and to calculate a binding affinity. (a) Expanded view of 1H, 15N HSQC spectra for ubiquitin alone (black) and with 2:1 (dark gray) or 1:1 (light gray) molar ratio of ubiquitin:hRpn13 (1–150). I44’s NH crosspeak shifts significantly, whereas that from Q2, which is not at the ubiquitin-binding interface, does not. (b) Chemical shift perturbation (CSP) values for ubiquitin NH signals when saturated with hRpn13 (1–150). Significantly shifted (labeled) and severely broadened (marked with asterisks) NH signals expose amino acids involved in binding. (c) Flow chart for modeling a ubiquitin:receptor structure with CSP values in HADDOCK. (d) CSPobs/CSPmax values are plotted against the molar ratio of Saccharomyces cerevisiae Rpn13 [R] to ubiquitin [U] and used in Matlab to calculate a binding constant of 65 μM. Each data line represents a specific amino acid, namely, F45 (black) and E72 (gray). (e) 1H, 13C HMQC spectra acquired on K48-linked diubiquitin with its proximal (left) or distal (right) moiety 13C labeled and no (black) or equimolar unlabeled hRpn13 (1–150) (gray). Significant shifting (indicated with arrows) is observed for I44 and A46 methyl groups (labeled) of the proximal but not the distal moiety (Panels (b) and (e) are reprinted by permission from Macmillan Publishers Ltd: Nature (51), copyright (2008)).
Record 2D heteronuclear correlation spectrum on free isotopically labeled ubiquitin. Example parameters for 1H, 15N (for 15N labeled ubiquitin) or 1H, 13C (for 13C labeled ubiquitin) correlation experiments are listed in Table 1 (see Note 10). Sample concentrations for labeled proteins typically range from 0.1 to 0.5 mM, although significantly lower concentrations can be used if coupled with longer experimental acquisition times with more scans per 2D increment.
Titrate in binding partner. The putative receptor is unlabeled and added from a concentrated stock to yield identical buffer conditions, as pH and salt concentration affect NMR signals. A recommended first titration point is 4:1 molar ratio of ubiquitin:receptor (see Note 11).
Record 2D heteronuclear correlation experiment on mixture. A heteronuclear correlation experiment is recorded and processed identically to step 1.
Repeat steps 2–3 until saturation. The putative receptor is added incrementally until no new spectral changes are observed or until it is at tenfold molar excess with no spectral changes from the free ubiquitin spectrum (see Note 12).
3.2.2. Using Heteronuclear Correlation Experiments to Map Binding Surfaces and Produce Model Complexed Structures
In the absence of structural changes, atoms with NMR signals that are most affected by the addition of a binding partner are typically at the interaction surface. Conversely, the mapping of affected signals to a localized surface suggests no structural rearrangements. Therefore, quantified comparison of chemical shift values for a protein in its free and bound state is often used to identify interaction surfaces. Such analysis is commonly done for 1H, 15N correlation experiments by using Eq. 1, in which CSP stands for chemical shift perturbation and ΔΔH and ΔΔN represent the change in the proton and nitrogen chemical shift values (in parts per million), respectively.
| (1) |
A CSP value is derived for all atoms observable in the NMR experiment and plotted, as demonstrated in Fig. 2b for ubiquitin binding to hRpn13.
When the structure and amide chemical shift assignments of the receptor is known, the titration can be done in both directions and the resulting CSP values for the two proteins used to model the complexed structure. Under such conditions, ambiguous interaction restraints (AIRs) can be generated from the CSP analysis for use in the docking program High Ambiguity Driven DOCKing (HADDOCK) (21), as described below and in the flow chart of Fig. 2c.
Define PDB files for ubiquitin and the receptor. PDB coordinates for the unbound structures of ubiquitin (PDB 1D3Z) and the receptor are inputted into HADDOCK through a user-friendly Web-based interface http://www.nmr.chem.uu.nl/haddock/.
Input active residues. Active residues can be defined as those with CSP values greater than or one standard deviation above the average value. In addition, the solvent accessibility of their main chain or sidechain atoms must be ≥50%.
Input passive residues. Passive residues neighbor or are proximal in space to active residues and have solvent accessibility ≥50%.
Input AIRs. AIRs are defined as ambiguous distance restraints and these enforce a maximum value of 2–3 Å between any atom from one protein’s active amino acids and an atom of its binding partner’s active or passive amino acids.
Input semiflexible segments within ubiquitin and the receptor. Semiflexible segments are defined as sequential spans of active and passive amino acids plus 1–2 additional amino acids on both sides.
-
Run HADDOCK. The information inputted in steps 1–5 is saved in a file named run.cns (see Note 13), which is used in HADDOCK to dock by rigid body energy minimization randomly oriented geometries of ubiquitin and the receptor into a complex that adheres to the AIRs. Lowest energy structures are subjected to semiflexible simulated annealing in torsion angle space followed by refinement in explicit water, and then sorted according to energy criteria.
In contrast to the methods discussed in Subheadings 3.3 and 3.4, this approach does not use distance-dependent information and therefore, it is necessary to use site-directed mutagenesis to confirm the resulting ubiquitin:receptor model structures. In particular, amino acids with sidechains predicted to form critical contacts should be substituted to disrupt binding. Effects can be tested by NMR-based titration experiments, as described in Subheading 3.2.3.
3.2.3. Using Heteronuclear Correlation Experiments for Binding Affinities
When NMR signals remain visible throughout a titration, the binding affinity can be obtained by plotting CSP values against their corresponding molar ratios, as shown in Fig. 2d. A protocol for 1:1 binding stoichiometry follows (see Note 14).
Acquire 1H, 15N HSQC experiments under identical conditions for 15N labeled ubiquitin alone and with unlabeled receptor at different molar ratio (such as 4:1, 2:1, 1:1, 1:2, 1:3, 1:4, 1:5, 1:6, 1:8, see Note 15), as described in Subheading 3.2.1.
Use Eq. 1 to calculate CSP values for ubiquitin signals at each molar ratio. Select those with significant chemical shift changes for the following analysis.
-
Plot the CSP values of selected ubiquitin signals against the molar ratio of receptor to ubiquitin and use Eq. 2 to calculate the dissociation constant Kd (see Note 16).
(2) In this equation, [R] and [U] are the concentration of receptor and ubiquitin, respectively. CSPobs and CSPmax are CSP values of selected ubiquitin signals at a corresponding [R]/[U] molar ratio and at saturation, respectively.
3.3. Paramagnetic Spin Labeling to Study Ubiquitin:Receptor Complexes
Spin labels containing unpaired electrons are introduced into proteins to cause distance-dependent paramagnetic relaxation enhancement (PRE) of NMR signals. PREs are observed in NMR spectra as reduced or lost signal and are proportional to r−6, where r is the distance between the observed atom and the unpaired electron center. They are readily quantified into distances on the 10–35 Å scale when using nitroxide spin labels and chelators of paramagnetic metal ions (22). To characterize the binding interface between ubiquitin and a receptor, 15N labeled ubiquitin is mixed with a receptor protein that is spin labeled at a specific site (see Note 17). PRE effects are observed in ubiquitin by a 1H, 15N correlation experiment, such as an HSQC experiment (Fig. 1a and Table 1, Experiment 1). An advantage of this approach is that it does not require the assignment of NMR signals for the receptor, although the complementary experiment can also be done if assignments are known; in this case, ubiquitin is spin labeled and the receptor is 15N labeled. Nitroxide spin label MTSL (23) is commonly used for spin labeling experiments, as it is small and offers thiol-specificity by forming disulfide bonds with cysteines (Fig. 3a). A disadvantage of MTSL is that it is labile with reducing agents (see Note 18). A protocol to define distances between ubiquitin and a receptor by MTSL labeling follows.
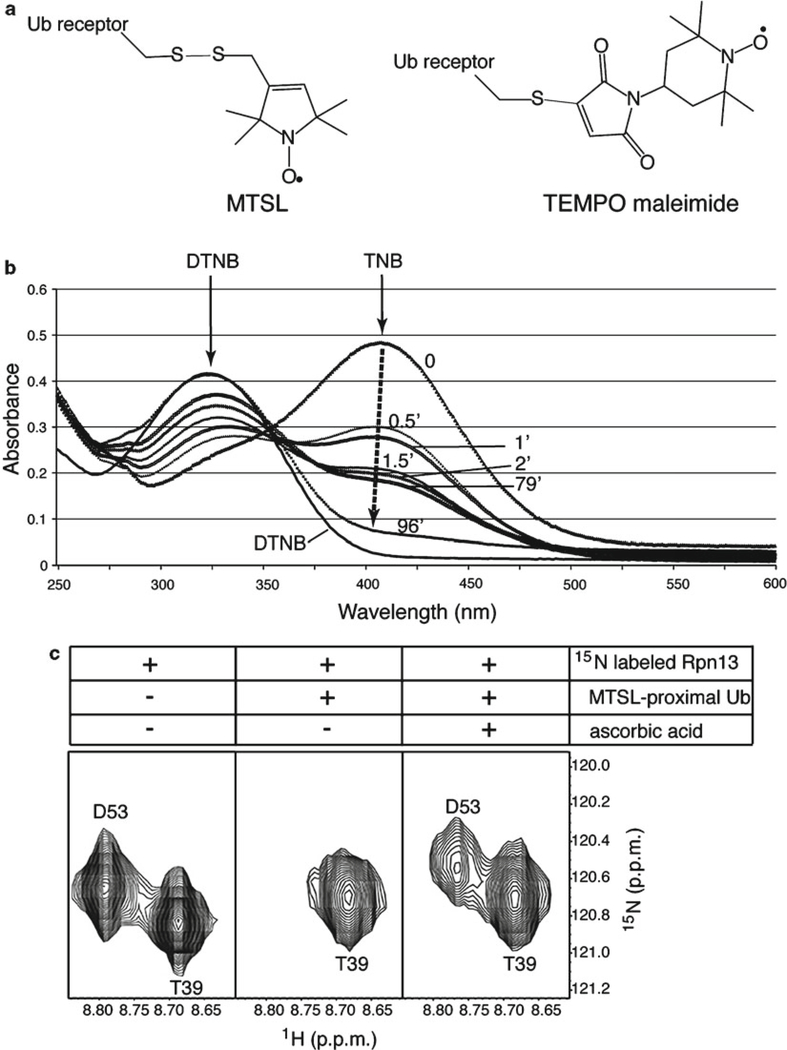
Fig. 3.
Use of spin labeling to characterize ubiquitin:receptor complexes. (a) Illustration of MTSL (left) or TEMPO maleimide (right) attached to a ubiquitin receptor by a cysteine thiol group via sulfur–sulfur or by carbon–sulfur bonds (gray), respectively. (b) Spin labeling reaction time course as measured by a DTNB assay. DTNB absorbs at 324 nm and reacts with free thiol groups to release 2-nitro-5-thiobenzoate (TNB), which absorbs at 412 nm and is not observed when spin labeling is complete (black dashed arrow). The plotted absorbance curves were recorded before adding MTSL (0 min) and after reacting it at equimolar quantity and 4°C in the dark for 0.5, 1, 1.5, 2, 79, and 96 min. (c) Expanded view of 1H, 15N HSQC spectra of hRpn13 (1–150) alone (left panel), with MTSL-labeled K48-linked diubiquitin G75C (middle panel), and with ascorbic acid added to the mixture (right panel). G75C-MTSL is on the proximal subunit and D53, but not T39, of hRpn13 experiences paramagnetic relaxation enhancement (middle), which is recovered by quenching (right) (Panel (c) is reprinted from ref. 39 with permission from Elsevier).
Check the structural integrity of the amino acid substituted receptor protein. Acquire 1H, 15N HSQC spectra on 15N labeled receptor without and with the needed mutations. Structural integrity is confirmed by few spectral changes. If assignments are known, only amino acids that are substituted themselves or proximal to those that are should shift.
Spin label the receptor protein. Dialyze an aliquot of purified receptor protein against 50 mM Tris–HCl, pH 7.6; no reducing agent should be present. Flush the reaction tube with N2 and then add three- to tenfold molar excess MTSL from a freshly prepared 150 mM acetonitrile stock. At room temperature and in the dark (since free MTSL is light sensitive), the reaction is typically completed within 10–30 min, depending on the initial protein concentration; incubation can also be done in the dark at 4°C overnight. A DTNB assay (24) is used to confirm that the labeling is complete (Fig. 3b). Following labeling, remove excess spin label by extensive dialysis at 4°C in the buffer to be used for the NMR experiment, such as Buffer 1. Mass spectrometry can be used to confirm successful labeling.
Acquire NMR data. Acquire a control 1H, 15N HSQC experiment on a sample of ≥0.3 mM 15N labeled ubiquitin. To this sample, add the MTSL-labeled receptor protein, preferably to saturation. Repeat the 1H, 15N HSQC experiment under identical conditions on the 15N-labeled ubiquitin:MTSL-labeled receptor complex; this spectrum contains MTSL in the paramagnetic (Ipara) state. Next, add fivefold molar excess ascorbic acid to the sample to quench the spin label; MTSL is reduced to its diamagnetic (Idia) state. After 1 h at 25°C, the quenched sample is subjected to another 1H, 15N HSQC experiment under identical conditions. Three such spectra are displayed for hRpn13 with K48-linked diubiquitin containing MTSL conjugated to a cysteine that has replaced G75 of its proximal subunit (Fig. 3c).
Calculate distances from PRE effects. Integrate the cross peaks in the two spectra recorded on 15N labeled ubiquitin:MTSL-labeled receptor from step 3. Plot the Ipara/ Idia ratio for the integrated values per amino acid residue. Typically, data analysis is done by establishing three groups of residues, defined as those with Ipara/Idia intensity ratios <15%, 15–85%, and >85%, and Eqs. 3 and 4 are used to assign distance ranges to each group (25–27).
| (3) |
| (4) |
The experimental measurement Ipara/Idia is used in Eq. 3 to obtain the amide 1H PRE rate Γ2, which is then used in Eq. 4 to define distances (r) between the unpaired electron center and amide 1H atoms. In Eq. 3, t is the time used in the experiment for the PRE to occur (~9 ms) (26, 27), R2,dia is the transverse relaxation rate of the amide 1H in its diamagnetic state, which can be estimated from the half-height line width (Δ𝜈FWHH) assuming Lorentzian line shapes (R2,dia = 𝜋Δ𝜈FWHH) ( 26, 28 ) (see Note 19). In Eq. 4, the constant K = 1.23 × 10−44 m6/s2 and 𝜔H is the proton Larmor frequency (3.77 × 109 and 4.40 × 109 rad/s for experiments recorded on 600 and 700 MHz spectrometers, respectively). The rotational correlation time (𝜏c) can be estimated from the molecular weight of the ubiquitin:receptor complex (Fig. 4), as r is not sensitive to errors in 𝜏c due to its (𝜏c)1/6-dependence (see Note 20). PREs can be used to qualitatively define interaction surfaces, but through this analysis, also to calculate the structure of ubiquitin:receptor complexes (Subheading 3.5).
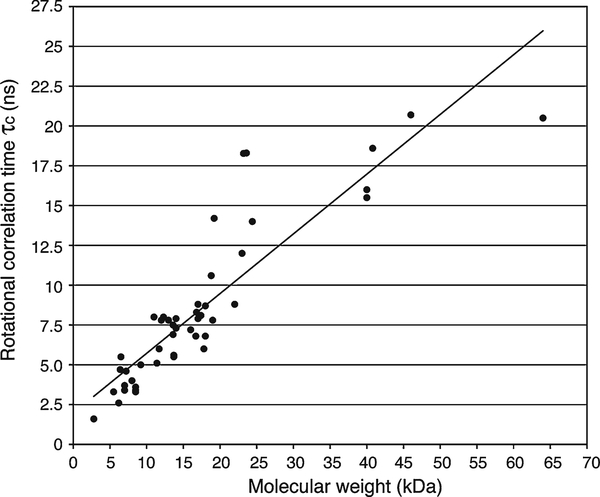
Fig. 4.
Experimentally derived relationship between rotational correlation time (𝜏c) and molecular weight for proteins and protein complexes ranging from 2.8 to 64 kDa. A total of 48 proteins or protein complexes are plotted according to their molecular weight in a manner similar to Fig. 1 of ref. 52, which plotted 39 of the values displayed here. The additional data points included are from the 13.6 kDa nitrogen regulatory protein C and its mutant (D86N/A89T) (25°C) (53), 23 kDa HIV-1 protease with a peptide substrate (20°C) (54), 23.6 kDa E. coli adenylate kinase (20°C) (55), 23 kDa Aquifex aeolicus adenylate kinase alone and with Mg·Ap5A (20°C) (55), 40 kDa N-terminal domain of enzyme I and the histidine-containing phosphocarrier protein complex (40°C) (56), 41 kDa maltose binding protein (37°C) (57), and 46 kDa OmpA transmembrane domain/micelles complex (58). Linear regression analysis was used to estimate a fit of y = 0.3751 x + 1.9684, where x is the molecular weight and y is the rotational correlation time.
3.4. Intermolecular Nuclear Overhauser Effects to Study Ubiquitin:Receptor Complexes
Intermolecular 1H–1H nuclear Overhauser effects (NOEs) between hydrogen atoms can be used to generate atomic resolution structures of ubiquitin:receptor complexes. Like the PRE effects described in Subheading 3.3, NOE interactions decay with an r−6 dependence, where r is the distance between the two 1H atoms.
They define shorter range distances however with an upper limit of ~6 Å. Unambiguous intermolecular NOEs between sidechain 1H atoms are obtained by 13C labeling only one of the components and then acquiring a 3D 13C-filtered, 13C-edited NOESY experiment (29, 30). This experiment detects NOE interactions only between 1H atoms attached to 13C and those attached to 12C, as interactions in which both 1H atoms are attached to 13C are filtered out. An example of the application of this approach to record intermolecular NOEs between ubiquitin and S5a is provided in Fig. 5a with experimental parameters listed in Experiment 3 of Table 1. Data analysis for this experiment requires knowledge of sidechain resonance assignments, which are readily available for ubiquitin (31) and can be obtained for a receptor by using modern NMR techniques (19, 20). A general protocol for implementing this class of experiment follows in which the receptor protein is 13C labeled and ubiquitin unlabeled. This experiment can also be acquired with 13C labeled ubiquitin and unlabeled receptor however.
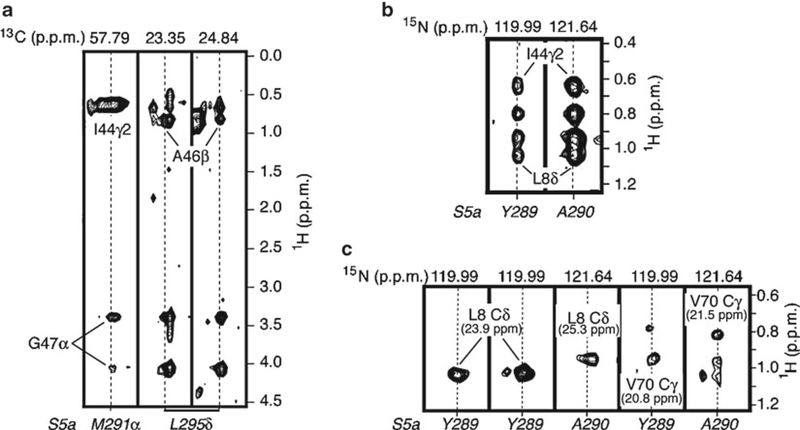
Fig. 5.
Intermolecular NOESY experiments to reveal ubiquitin-receptor contacts within 6 Å. (a) Selected strips from a 3D 13C-filtered, 13C-edited NOESY spectrum recorded on 13C-labeled S5a (196–306) and unlabeled ubiquitin. (b) Selected 15N planes from a 3D 15N-dispersed NOESY spectrum recorded on 2H, 15N-labeled S5a (196–306) and unlabeled ubiquitin. (c) Selected 13C planes from a 4D 15N-edited, 13C-dispersed NOESY on 2H, 15N -labeled S5a (196–306) and 13C-labeled ubiquitin. Atom labels at the bottom of the strips (in italics belong to S5a, whereas all others belong to ubiquitin. Parameters) for these experiments are listed in Table 1. In all cases, the concentration of S5a and ubiquitin is 0.5 mM and 2.0 mM, respectively (Reprinted from ref. 33 with permission from Elsevier).
Produce required sample. Prepare ~0.5 mM complexed sample dissolved in 99.96% D2O (see Subheading 3.1) with the 13C labeled receptor saturated with unlabeled ubiquitin. Validate the integrity of the labeled receptor alone and with ubiquitin by 1H, 13C HSQC experiments (see Subheading 3.2). Spectral changes should be observed upon ubiquitin addition and the bound state NMR signals must be observable.
Check 2D planes. Acquire each 2D plane of the 13C-filtered, 13C-edited NOESY (100 ms mixing time) experiment. The 1H, 13C plane should resemble the bound state spectrum, as described in step 1. The 1H, 1H spectrum should show weak off diagonal cross peaks and the diagonal signals should be filtered out. Acquiring the 2D planes takes ~1 h, although higher signal-to-noise can be obtained with longer experimental times. The 2D versions are used to optimize parameters for the 3D experiment.
Acquire 3D experiment. Acquire the 3D 13C-filtered, 13C-edited NOESY (100 ms mixing time) experiment. For large proteins, higher sensitivity is achieved by using an HMQC module for 13C selection. An example of typical parameters for this experiment is provided in Experiment 3 of Table 1 and the experimental time is ~5 days.
Validate samples after 3D experiment. After the 3D experiment is finished, a 1H, 13C HSQC experiment and SDS-PAGE (on ~2 μL of NMR sample) are used to check the sample integrity.
If only the amide resonance assignments are available for the receptor, intermolecular NOEs can be obtained by acquiring an 15N-dispersed NOESY experiment on 100% 2H- and 15N labeled receptor mixed with unlabeled ubiquitin (32), as demonstrated for S5a:ubiquitin in Fig. 5b. This experiment offers less NOE interactions compared to the 13C half-filtered experiment mentioned above and is generally not sensitive enough for larger complexes >20 kDa. A general protocol follows.
Produce required sample. 2H, 15N labeled receptor is prepared as described in Subheading 3.1 and its integrity is validated by a 1H, 15N HSQC spectrum, as described in Subheading 3.2. Complexed sample of 0.5 mM with the receptor saturated is checked by a 1H, 15N HSQC experiment. Spectral changes and the bound state must be observed.
Check 2D planes. Acquire the 2D 1H, 1H and 1H, 15N planes of the 15N-edited NOESY (200 ms mixing time) experiment. The 1H, 15N plane should resemble the bound state spectrum of step 1 and weak NOE interactions should be apparent in the 1H, 1H plane. Note that NOEs to H2O are observable, as are intramolecular amide-to-amide NOEs. The 2D experiments are used to optimize 3D parameters.
Acquire 3D experiment. Acquire a 3D 15N-edited NOESY (200 ms mixing time) experiment. Example parameters are provided in Experiment 4 of Table 1.
Validate sample after 3D experiment. A 1H, 15N HSQC experiment and SDS-PAGE are used to check the integrity of the NMR sample, as described for the 13C-filtered, 13C-edited NOESY experiment.
The above NOESY experiment can also be done with 13C editing of the ubiquitin sidechain 1H resonances to help with assigning NMR signals to ubiquitin atoms. A 13C dispersed, 15N-edited NOESY experiment is acquired on a sample containing 15N, 2H labeled receptor and 13C labeled ubiquitin (33), as demonstrated for S5a:ubiquitin in Fig. 5c with parameters listed in Experiment 5 of Table 1. Additional experimental time is required to label the NMR signal with the 13C information, which causes the experiment to be less sensitive and to have fewer intermolecular NOEs.
Intermolecular NOEs between a receptor and ubiquitin moieties within a ubiquitin chain can also be obtained by using the NOESY experiments described above. A ubiquitin moiety can be selectively labeled during synthesis and 2H labeling used to block signals from certain moieties. For example, specific intermolecular NOEs can be obtained between a receptor and the distal subunit of diubiquitin by acquiring a 13C-filtered, 13C-edited NOESY experiment on 13C labeled receptor mixed with diubiquitin in which the proximal subunit is 100% 2H labeled and the distal subunit is unlabeled (39 ).
3.5. Structure Calculations of Ubiquitin:Receptor Complexes
The structure of ubiquitin:receptor complexes can be calculated by using NMR-derived restraints in structural calculation programs, such as Xplor-NIH (34, 35) or CNS (36). We provide two examples; the first, simpler, rigid body docking approach can be used when the structure of the receptor is available and no significant structural rearrangements occur upon binding for either protein. The second approach is necessary when the proteins structures must also be derived during the docking. Both use scripts available in Xplor-NIH.
Generate protein structural files (PSFs) for ubiquitin and the receptor. Use the script “mkpsf_both.inp” (provided in the Xplor-NIH directory eginput/prot-prot) to generate a multi-segment PSF (see Note 21) for ubiquitin and the receptor from their amino acid sequences (Fig. 6, Example 1).
Prepare input restraint files. Subheading 3.3 describes how to convert PRE effects into distances between 1H atoms and a spin label (see Note 22). NOE interactions are converted into distances by using their r−6 dependence (see Subheading 3.4) (see Note 23). Figure 6, Example 2 lists examples of input restraint files.
Setup scripts and run program. The script “rigid_min.inp” docks ubiquitin and the receptor together as rigid bodies and performs a series of energy minimization steps involving the experimentally derived distances and van der Waals repulsion to generate single starting structure for “sa_cross_tor.inp.” “rigid_min.inp” is edited to define the PSF generated in step 1 and the starting PDB files for the free proteins, which should be saved in the same directory as the PSF. “sa_cross_tor.inp” is edited to define experimental restraints, the PSF, and the number of complexed structures generated by energy minimization and simulated annealing. This script keeps the backbone angles for most amino acids rigid during the docking, while allowing those at the interface greater mobility. In its final stage, 40,000 steps of energy minimization are performed and final structures written out as coordinate files (Fig. 6, Example 4) (see Note 24).
Analyses and validation. The events of step 3 are listed in an output file that tracks the progress of the script. This file also lists all violations in the final outputted coordinate file for each structure calculated. Each violation needs to be carefully checked against the experimental data, as they invariably arise from human error. After fixing any errors, steps 2 and 3 are repeated. Final structures should have no experimentally derived distance violation above 0.3 Å or dihedral angle violation above 5°.
Fig. 6.
Example flow chart and scripts for calculating ubiquitin:receptor complexes with Xplor-NIH starting from randomly coiled structures. The number of structures calculated, temperatures used, and the number of steps and step size can all be altered as desired. The listed values have worked well for ubiquitin:receptor complexes of ~25 kDa in size. Example excerpts from PSF (1) and restraints (2) files, a simulated annealing script (3), and an output coordinate file (4) are provided.
A more complicated protocol to calculate the three-dimensional structure of a ubiquitin:receptor complex by using NMR-based restraints in Xplor-NIH follows. In this case, the structures of the proteins are calculated during the docking.
Generate PSF and coordinate input files for ubiquitin and the receptor (Fig. 6, step 1). Use the script “seq2psf” provided in the Xplor-NIH package to generate separate PSFs (Fig. 6, Example 1) for ubiquitin and the receptor from their amino acid sequences. Use “generate_template.inp” to generate separate template coordinate files for ubiquitin and the receptor (see Note 25). Separate segment identifiers need to be assigned explicitly in the PSFs and template coordinate files. Generate a merged template coordinate file from the individual files (see Note 26).
Prepare input restraint files (Fig. 6, step 2). The experimentally derived intermolecular distance restraint files are generated as described for the previous method (Fig. 6, Example 2). Intramolecular restraints are generated experimentally by using intramolecular NOESY and PRE data in a manner analogous to the intermolecular restraints (see Note 27). In the absence of structural changes in ubiquitin, the available intramolecular restraints for free ubiquitin can be used (PDB code 1D3Z) (31).
Setup scripts and run program (Fig. 6, steps 3–7). A simulated annealing script for protein ligand docking in Xplor-NIH is edited to define the input files created in steps 1–2 and the number of structures calculated (Fig. 6, Example 3). Structure calculations are performed with a Linux operating system and Xplor-NIH version 2.24 (34, 35) in five stages, as illustrated in Fig. 6. Randomly coiled starting structures are generated and subjected to energy minimization to ensure full spatial sampling and appropriate coordinate geometry. The structures are next confined according to the inputted restraint files by subjecting them to 15,000 simulated annealing steps of 0.005 ps at 3,000 K, followed by 10,000 cooling steps of 0.005 ps. 2,000 steps of energy minimization is then applied and the final structures written out as coordinate files (Fig. 6, Example 4).
Analyses and Validation (Fig. 6, step 8). This stage mimics that described above for the rigid body docking protocol. Typically, 10–15 NOEs per amino acid residue are required for NMR-based structure calculations when starting from randomly coiled structures.
Acknowledgments
We are grateful to Leah Randles and Hiroshi Matsuo for their critical reading of this manuscript. Research in the K.J.W. laboratory is supported by the National Institutes of Health (CA097004, CA117888, and CA136472) and the American Cancer Society (RSG-07–186-01-GMC).
Footnotes
Some proteins require only 2 g/L of glucose, which can reduce costs when isotopically labeled glucose is required. Trial mini-inductions at the 10 mL scale can be performed to determine glucose requirements.
All components for 99.9% deuterated M9 minimal media should be anhydrous. Autoclave and dry all glassware before use.
For experiments involving amide protons, a pH≤7 is recommended to minimize their exchange with water.
Larger systems (>20 kDa) typically require a cryogenically cooled probe and therefore salt concentrations <100 mM. Higher salt concentrations can be used with conventional or salt-tolerant cryogenically cooled probes.
Additives like detergents or metal ions can be included in the buffer used for NMR experiments when necessary to stabilize the sample and 2H labeled options exist for many reagents to minimize buffer peaks in NMR spectra.
A variety of NMR tube options exist, some of which are optimized for high salt, low volume, or low sample concentration.
To produce fully 2H labeled protein, D-glucose-1,2,3,4,5,6,6d7 (97 atom% D) is used in the final 1 L growth media and cells are adapted incrementally to D2O (32). Typically, 100 μL of an LB starter culture is used to inoculate a 10 mL M9 culture that contains 50% D2 O/50% H2O. In the ideal case, 100 μL of this culture can be used to inoculate a 10 mL M9 culture containing 99.9% D2O, which is then used to inoculate a 1 L culture containing 2H labeled glucose and 99.9% D2O. Each of the 10 mL cultures is grown to confluency before stepping up to the next culture. For most proteins, growth in D2O is significantly slower compared to H2O, and antibiotic and Thiamine–HCl, which break down, need to be added every 12 h. Some proteins require 10 mL M9 cultures containing 50 or 70% D2O be used to inoculate the 1 L culture containing 99.9% D2O, whereas others are unable to be expressed at all in 99.9% deuterated media. Small-scale tests are recommended prior to large-scale productions. Following expression, cells are pelleted and their lysis and the protein purification are done in buffers containing H2O.
The temperature of NMR experiments varies according to sample needs and most ubiquitin systems are studied between 20 and 35°C.
An additional advantage of using ubiquitin as the labeled protein exists for receptors that cannot be expressed and purified from E. coli.
In most cases, 15N labeling is used as it readily monitors all amino acids except proline and with the exception of methyl groups, generally exhibits less spectral overlap. The signal from methyl groups is well dispersed and strongest and therefore, for large systems, such as those involving ubiquitin chains, it is generally advantageous to use 13C labeling and 1H, 13C correlation experiments. The individual moieties of ubiquitin chains can be specifically isotopically labeled to reduce spectral crowding (Fig. 2e). Highest sensitivity is achieved for 1H, 13C 2D correlation experiments when samples are dissolved in 99.96% D2O.
It is not necessary to start with a molar ratio of 4:1, but the incremental addition of receptor to ubiquitin enables the NMR signals to be readily followed from their free to their bound states.
Since they occur at relatively high protein concentration (μM or higher) and sample amino acids across the whole protein sequence, weak binding events are readily detectable by 2D correlation experiments, even when the putative binding partner is only twofold in molar excess over the labeled one. When no spectral changes are observable upon the titration of one protein into another, the two proteins do not interact under the experimental conditions used.
Default parameters, such as the number of structures generated, can be changed in the run.cns file through the Web-based interface.
This approach can be also with 15N labeled receptor and unlabeled ubiquitin and equations exist for complexes with 2:1 binding stoichiometry (37).
During the titration, unlabeled receptor should be added in small aliquots (≤10 μL) from a concentrated stock. If a large volume of receptor must be added (due to receptor concentration), then the NMR sample should be concentrated to keep the ubiquitin concentration constant.
The CSP values are used with the corresponding concentrations of receptor and ubiquitin to fit Eq. 2 in a mathematical program, such as Matlab v. 7.2.
Most commonly, cysteine residues are used to introduce spin labels into proteins. Specificity is achieved by the alanine substitution of all native cysteines that are not used for labeling and the introduction of cysteine at desired labeling sites. Ubiquitin has no native cysteines, but they have been introduced into it and its polymers for spin labeling (37–39). It is important that the spin label be proximal to, but not within, the contact surface. Often data from multiple sites is used in structure calculations and labeling that alters binding properties is not used; PRE data is obtained by using 1H, 15N correlation experiments, which readily reveal if binding or protein structure has been compromised, see Subheading 3.2.
When the receptor requires a reducing agent for stability, the bulkier spin label TEMPO maleimide (40) can be used, as it forms a carbon–sulfur bond with cysteine (Fig. 3a). Maleimide can react with primary amines or be hydrolyzed at pH values >7.5 (41); however, NMR experiments are usually conducted at lower pH.
More vigorous, but significantly more complicated methods exist to obtain R2,dia as described in ref. 42. A series of 2D experiments with varying relaxation delay is acquired and the resulting data fit to a single exponential decay function.
τc for ubiquitin is ~4 ns ( 43).
PSFs contain atomic details and covalent geometry information.
For PRE-derived distances, atomic and geometric details for the spin label are included in the PSF and template coordinate files, which is achieved by editing the sequence file to include cysteine conjugated to the spin label, as defined in the Xplor-NIH topology file “protein.top” (such as for MTSL). For spin labels not already in the Xplor-NIH topology file, a topology file can be generated, for example with the Web-based program PRODRG http://davapc1.bioch.dundee.ac.uk/prodrg/ (44).
It is convenient to calibrate the intermolecular NOEs by assigning a distance of 6 Å to the peak volume of the weakest crosspeaks.
Each outputted coordinate file lists that structure’s energy contribution profile for experimentally derived restraints (NOE- or PRE-based distances, hydrogen bond distances, and dihedral angles) as well as for the target atom and bond geometries defined within the Xplor-NIH protein parameter file “protein.par.” Root-mean-square deviation (RMSD) values between ideal and calculated atom and bond geometries are also listed, along with the number of distance and dihedral angle restraints violated above thresholds defined in the script.
Other scripts included in the Xplor-NIH package, such as “protG_mkpsf.inp,” can be edited to generate PSF and template files from a protein’s amino acid sequence.
Use a protein structure viewer package, such as INSIGHT II (Accelrys) or Swiss PDB viewer, to merge the individual template coordinate files of ubiquitin and the receptor into one saved coordinate file. This new file is used by “generate.inp” to create a new PSF file, which is next used by “generate_template.inp” to create a merged template coordinate file.
The three-dimensional structure of the receptor and ubiquitin can be defined by separate restraint files that list distance restraints derived by intramolecular NOE interactions and PRE effects, hydrogen bond restraints based on secondary structure information, and 𝜑 and 𝜙 dihedral angle restraints. Distance restraints for intramolecular PRE data can be generated as described in Note 22 and Subheading 3.3 for intermolecular PREs. In this case, the spin-labeled protein is 15N labeled while the binding partner is unlabeled. NOE-based restraints can be derived by using CARA (Computer-Aided Resonance Assignment; Diss. ETH No. 15947) to calibrate the NOESY spectra and prepare an assignment file, followed by ATNOS (automated NOESY peak picking) (45) and CANDID (combined automated NOE assignment and structure determination module) (46) to pick and assign NOESY peaks and convert them to distance restraints. It is worth noting that this assignment process for NOEs often requires multiple cycles of user intervention and refinement. Secondary structural elements are defined by comparing the assigned chemical shift values of carbonyl and Cα atoms to those of randomly coiled structures; these values are larger in helical structures and smaller for amino acids in β-strands (47, 48). NOE interactions also define secondary structure. For example, NOEs from Hα of amino acid i to NH of amino acid i + 4 and to Hβ of amino acid i + 3 are observed in α-helices, whereas unique NOE interactions are observed between Hα atoms of juxtaposed antiparallel β strands (19). Hydrogen bond restraint files are generated from secondary structure assignments, and confine the distance from the acceptor oxygen to the donor hydrogen and to the donor nitrogen as 1.8–2.0 Å and 2.7–3.0 Å, respectively (49). 𝜑 and 𝜙 dihedral angles are assigned via the user-friendly program TALOS+ (50), which uses the chemical shift assignment for the main chain N, NH, Hα, Cα, carbonyl, and sidechain Cβ.
References
- 1.Liu F, Walters KJ (2010) Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci 35 :352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains – from structures to functions. Nat. Rev Mol Cell Biol 10: 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walters KJ, Ferentz AE, Hare BJ et al. (2001) Characterizing protein-protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods Enzymol 339:238–258. [DOI] [PubMed] [Google Scholar]
- 4.Nietlispach D, Mott HR, Stott KM et al. (2004) Structure determination of protein complexes by NMR. Methods Mol Biol 278:255–288. [DOI] [PubMed] [Google Scholar]
- 5.Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20 S proteasome by NMR. Nature 445:618–622. [DOI] [PubMed] [Google Scholar]
- 6.Religa TL, Sprangers R, Kay LE (2010) Dynamic regulation of archaeal proteasome gate opening as studied by TROSY NMR. Science 328:98–102. [DOI] [PubMed] [Google Scholar]
- 7.Ruschak AM, Religa TL, Breuer S et al. (2010) The proteasome antechamber maintains substrates in an unfolded state. Nature 467:868–871. [DOI] [PubMed] [Google Scholar]
- 8.Raasi S, Pickart CM (2005) Ubiquitin chain synthesis. Methods Mol Biol 301:47–55. [DOI] [PubMed] [Google Scholar]
- 9.Pickart CM, Raasi S (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol 399:21–36. [DOI] [PubMed] [Google Scholar]
- 10.Komander D, Lord CJ, Scheel H et al. (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell 29:451–464. [DOI] [PubMed] [Google Scholar]
- 11.Bremm A, Freund SM, Komander D (2010) Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 17:939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD (2008) Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem 283:19581–19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Virdee S, Ye Y, Nguyen DP et al. (2010) Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat Chem Biol 6:750–757. [DOI] [PubMed] [Google Scholar]
- 14.Eger S, Scheffner M, Marx A, Rubini M (2010) Synthesis of Defined Ubiquitin Dimers. J Am Chem Soc 132:16337–16339. [DOI] [PubMed] [Google Scholar]
- 15.Torizawa T, Shimizu M, Taoka M et al. (2004) Efficient production of isotopically labeled proteins by cell-free synthesis: a practical protocol. J. Biomol. NMR 30:311–325. [DOI] [PubMed] [Google Scholar]
- 16.Pickford AR, O’Leary JM (2004) Isotopic labeling of recombinant proteins from the methylotrophic yeast Pichia pastoris. Methods Mol Biol 278:17–33. [DOI] [PubMed] [Google Scholar]
- 17.Sugiki T, Shimada I, Takahashi H (2008) Stable isotope labeling of protein by Kluyveromyces lactis for NMR study. J Biomol NMR 42: 159–162. [DOI] [PubMed] [Google Scholar]
- 18.Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wüthrich K (1986) NMR of Proteins and Nucleic Acids, Wiley, New York. [Google Scholar]
- 20.Cavanagh J, Fairbrother WJ, Palmer AG et al. (2006) Protein NMR Spectroscopy: Principles & Practice (Second Edition), Academic Press Inc., San Diego. [Google Scholar]
- 21.Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737. [DOI] [PubMed] [Google Scholar]
- 22.Clore GM, Iwahara J (2009) Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 109:4108–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berliner LJ, Grunwald J, Hankovszky HO, Hideg K (1982) A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal Biochem 119:450–455. [DOI] [PubMed] [Google Scholar]
- 24.Riddles PW, Blakeley RL, Zerner B (1979) Ellman’s reagent: 5,5 - dithiobis(2-nitrobenzoic acid) – a reexamination. Anal Biochem 94:75–81. [DOI] [PubMed] [Google Scholar]
- 25.Solomon I, Bloembergen N (1956) Nuclear magnetic interactions in the HF molecule. J Chem Phys 25:261–266. [Google Scholar]
- 26.Battiste JL, Wagner G (2000) Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear Overhauser effect data. Biochemistry 39:5355–5365. [DOI] [PubMed] [Google Scholar]
- 27.Iwahara J, Tang C, Clore GM (2007) Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson 184:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang B, Bushweller JH, Tamm LK (2006) Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J Am Chem Soc 128:4389–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wider G, Weber C, Wüthrich K (1991) Proton-Proton Overhauser Effects of Receptor-Bound Cyclosporin A Observed with the Use of a Heteronuclear-Resolved Half-Filter Experiment. J Am Chem Soc 113:4676–4678. [Google Scholar]
- 30.Lee W, Revington MJ, Arrowsmith C, Kay LE (1994) A pulsed field gradient isotope-filtered 3D 13 C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett 350:87–90. [DOI] [PubMed] [Google Scholar]
- 31.Cornilescu G, Marquardt JL, Ottiger M, Bax A (1998) Validation of Protein Structure from Anisotropic Carbonyl Chemical Shifts in a Dilute Liquid Crystalline Phase. J Am Chem Soc 120:6836–6837. [Google Scholar]
- 32.Walters KJ, Matsuo H, Wagner G (1997) A simple method to distinguish intermonomer nuclear Overhauser effects in homodimeric proteins with C2 symmetry. J Am Chem Soc 119:5958–5959. [Google Scholar]
- 33.Wang Q, Young P, Walters KJ (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol 348:727–739. [DOI] [PubMed] [Google Scholar]
- 34.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160:65–73. [DOI] [PubMed] [Google Scholar]
- 35.Schwieters CD, Kuszewski JJ, Clore GM (2006) Using Xplor-NIH for NMR molecular structure determination. Progr NMR Spectroscopy 48:47–62. [Google Scholar]
- 36.Brunger AT, Adams PD, Clore GM et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54:905–921. [DOI] [PubMed] [Google Scholar]
- 37.Varadan R, Assfalg M, Haririnia A et al. (2004) Solution conformation of Lys63-linked diubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279:7055–7063. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Raasi S, Fushman D (2008) Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol 377:162–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang N, Wang Q, Ehlinger A et al. (2009) Structure of the S5a:K48-linked diubiquitin complex and its interactions with Rpn13. Mol Cell 35:280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Griffith OH, McConnell HM (1966) A nitroxide-maleimide spin label. Proc Natl Acad Sci USA 55:8–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klare JP, Steinhoff HJ (2009) Spin labeling EPR. Photosynth Res 102:377–390. [DOI] [PubMed] [Google Scholar]
- 42.Donaldson LW, Skrynnikov NR, Choy WY et al. (2001) Structural characterization of proteins with an attached ATCUN motif by paramagnetic relaxation enhancement NMR spectroscopy. J Am Chem Soc 123:9843–9847. [DOI] [PubMed] [Google Scholar]
- 43.Tjandra N, Feller SE, Pastor RW, Bax A (1995) Rotational Diffusion Anisotropy of Human Ubiquitin from 15 N NMR Relaxation. J Am Chem Soc 117:12562–12566. [Google Scholar]
- 44.Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. [DOI] [PubMed] [Google Scholar]
- 45.Herrmann T, Guntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE-identifi cation in the NOESY spectra using the new software ATNOS. J Biomol NMR 24:171–189. [DOI] [PubMed] [Google Scholar]
- 46.Herrmann T, Guntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227. [DOI] [PubMed] [Google Scholar]
- 47.Wishart DS, Bigam CG, Holm A et al. (1995) 1 H, 13 C and 15 N random coil NMR chemical shifts of the common amino acids. J Biomol NMR 5:67–81. [DOI] [PubMed] [Google Scholar]
- 48.Schwarzinger S, Kroon GJ, Foss TR et al. (2001) Sequence-dependent correction of random coil NMR chemical shifts. J Am Chem Soc 123:2970–2978. [DOI] [PubMed] [Google Scholar]
- 49.Guntert P (1997) Calculating protein structures from NMR data. Methods Mol Biol 60:157–194. [DOI] [PubMed] [Google Scholar]
- 50.Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schreiner P, Chen X, Husnjak K et al. (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner G (1997) An account of NMR in structural biology. Nat Struct Biol 4 Suppl:841–844. [PubMed] [Google Scholar]
- 53.Volkman BF, Lipson D, Wemmer DE, Kern D (2001) Two-state allosteric behavior in a single-domain signaling protein. Science 291: 2429–2433. [DOI] [PubMed] [Google Scholar]
- 54.Katoh E, Louis JM, Yamazaki T et al. (2003) A solution NMR study of the binding kinetics and the internal dynamics of an HIV-1 protease-substrate complex. Protein Sci 12:1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henzler-Wildman KA, Lei M, Thai V et al. (2007) A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature 450:913–916. [DOI] [PubMed] [Google Scholar]
- 56.Garrett DS, Seok YJ, Peterkofsky A et al. (1999) Solution structure of the 40,000 Mr phosphoryl transfer complex between the N-terminal domain of enzyme I and HPr. Nat Struct Biol 6:166–173. [DOI] [PubMed] [Google Scholar]
- 57.Hwang PM, Skrynnikov NR, Kay LE (2001) Domain orientation in beta-cyclodextrin-loaded maltose binding protein: diffusion anisotropy measurements confirm the results of a dipolar coupling study. J Biomol NMR 20:83–88. [DOI] [PubMed] [Google Scholar]
- 58.Tamm LK, Abildgaard F, Arora A et al. (2003) Structure, dynamics and function of the outer membrane protein A (OmpA) and influenza hemagglutinin fusion domain in detergent micelles by solution NMR. FEBS Lett 555:139–143. [DOI] [PubMed] [Google Scholar]