Figure 2. Comparing the Composition of Small RNAs in Activated T Cells, EVs, and Aggregates.
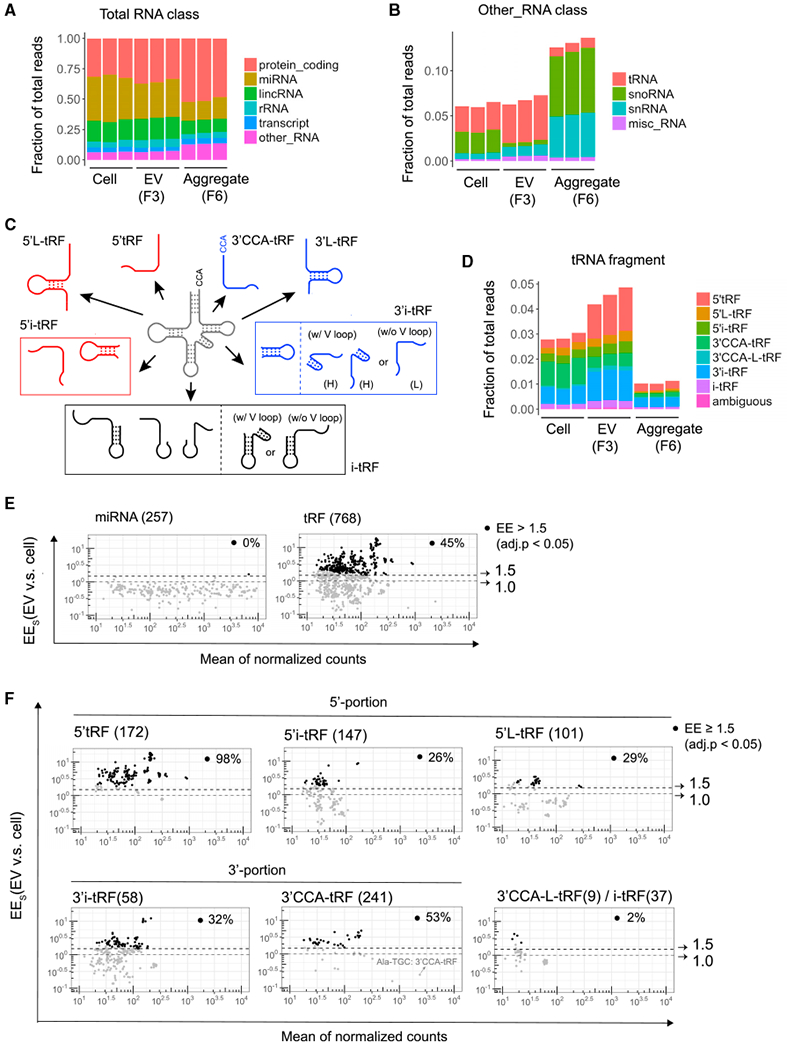
(A) Relative composition of mapped reads in small RNA libraries. RNA classification was based on the biotypes of Ensembl gene and transcript annotation. The “transcript” RNA class is the union of different biotypes, including processed transcript, antisense, and pseudogene.
(B) Sub-classification of reads shown as “other_RNA” in (A) into tRNA, snoRNA, snRNA, and misc_RNA.
(C) Pictorial summary of the 7 categories of tRFs that can arise from mature tRNAs. 3′i-tRF were further classified as hairpin (H) or linear (L) to indicate the presence or absence of tRNA variable loop-derived hairpin structures, respectively.
(D) Classification of reads in the 7 tRF classes shown in (C) and rare tRFs of ambiguous classification.
(E and F) MA plots comparing EV enrichment (EEs) (ratio of reads in EVs versus cells) and mean of normalized counts per million for all 257 detected miRNAs (E, left), all 768 detected tRFs (E, right), and subsets of tRFs in the indicated classes (F). Dotted lines and arrows indicate thresholds at 1.0 (no enrichment) and 1.5 EE. Black circles (·) mark RNA species significantly enriched in exosomes (≥ 1.5-fold; adjusted p < 0.05), and the percentage of significantly enriched RNA species among each class is indicated in each plot.
