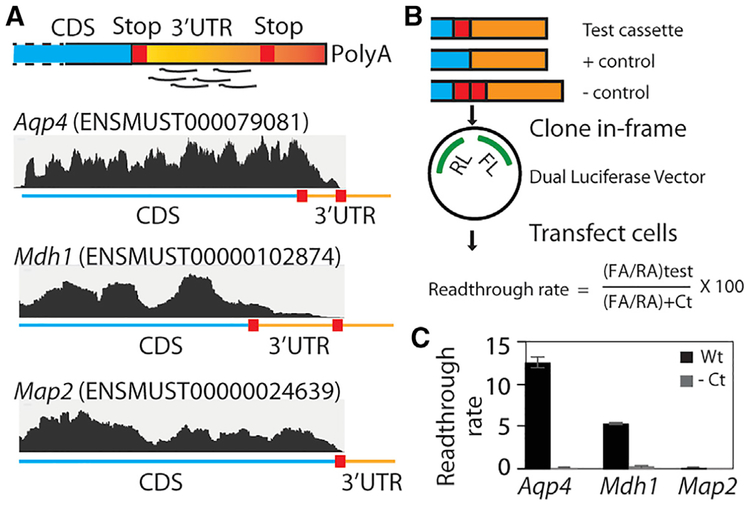
Figure 4. RF of Cycloheximide-Treated Neuron-Glia Culture Reveals Stop Codon Readthrough In Vitro.
(A) A schematic of footprint mapping and genome browser tracts with examples of readthrough are shown. Aqp4 and Mdh1 display footprints mapping to the readthrough region, whereas Map2 does not.
(B) Dual-luciferase assay assessing the permissiveness of sequences for readthrough. A cassette spanning the distal CDS and the readthrough region is cloned between the Renilla luciferase (RL) and firefly luciferase (FL) such that the latter is expressed in transfected cells only if ribosomes read past the cassette. The stop codon is mutated to a sense codon in the positive control, whereas an extra stop codon is added in the negative control.
(C) Dual-luciferase assay shows that Aqp4 and Mdh1 undergo readthrough at the rates of 12.5% and 5.5%, respectively, and that Map2 does not undergo readthrough. Readthrough rate is calculated as shown in (B) (n = 3; error bar, SEM).
CDS, coding sequence; RA, Renilla activity; FA, firefly activity; Ct, control. Also see Table S3 and Figure S3.