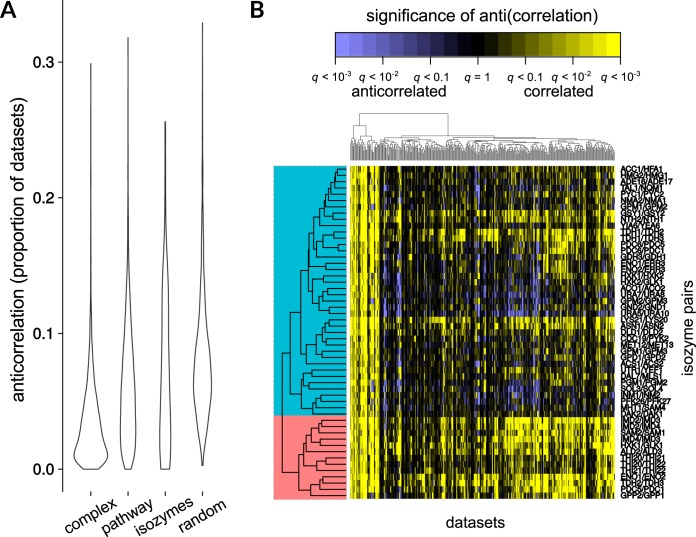
FIG 1.
Many isozyme pairs are differentially expressed. (A) Box plots of anticorrelation among isozyme pairs compared with (i) members of the same protein complex, (ii) members of the same metabolic pathway, and (iii) random gene pairs. Isozyme pairs are more likely to show differential expression than genes in the same complex (Bonferroni-Holm-corrected 2-sided Wilcox test P value, 1.0 × 10−4) but less likely than random genes (Holm-corrected P = 0.031). (B) Isozyme pairs separate into two broad categories, depending on how often they are anticorrelated. The matrix displayed shows the correlation (yellow) or anticorrelation (blue) of isozyme pairs (rows) for every data set (columns) in the compendium. Intensity data correspond to the significance of the (anti)correlation (q value). Hierarchical clustering performed using an uncentered Pearson’s correlation reveals two main clusters of isozyme pairs; a minority are strongly correlated over most of the compendium, while the majority show condition-dependent anticorrelation.