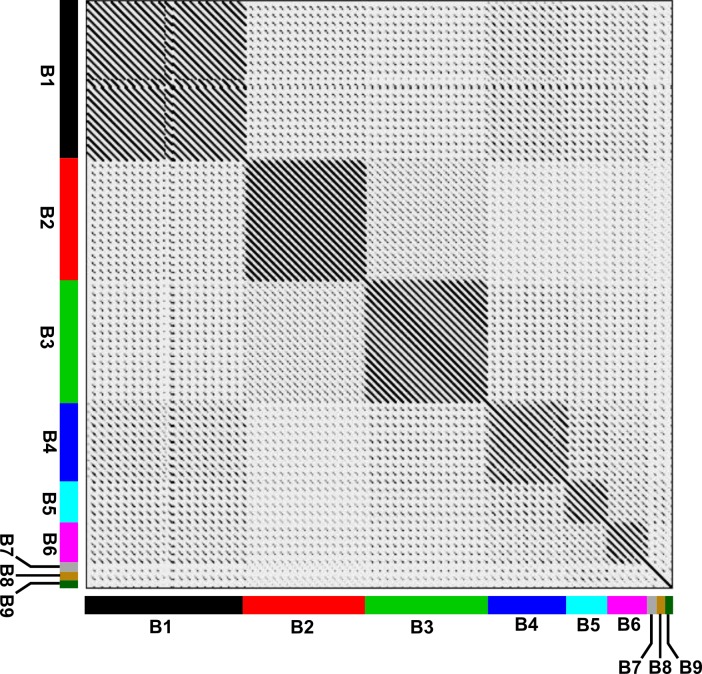
Fig 6. Dotplot comparison of TMPs from 75 mycobacteriophages of cluster B using Gepard.
The different subclusters are displayed in colored bars in both axes.Of note, the analysis of the aminoacid sequence of CRB2 TMP revealed a fragment encompassing 69 residues which displayed high homology with a family of proteins known as Resuscitation Promoting Factors (Rpf) as shown in S2 Fig [3]. A comparable domain was also detected in phages Saguaro and Thonko (S2 Fig). Of note, this domain has not been previously described in phages of the B cluster although it has been reported for some phages belonging to different clusters, such as Barnyard and Giles [3,35]. It has been postulated that those proteins are involved in activating or “waking-up” dormant host cells prior infection in the case of phages and in peptidoglycan recycling as a metabolic alarm for quiescent M. tuberculosis cells and in general for mycobacterial cells. This analysis suggests that CRB2, Thonko and Saguaro may be able to infect M. smegmatis cells in stationary phase, a hypothesis yet to be tested.