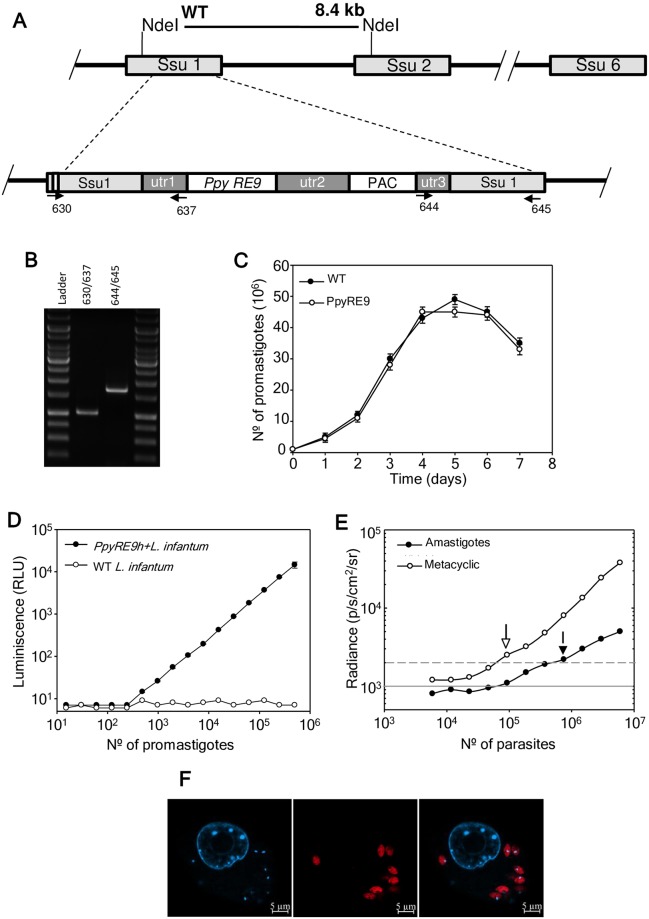
Fig 1. Generation of a L. infantum stably expressing PpyRE9h red-shifted luciferase.
A) Scheme of the structure of the 18S rRNA locus on wild type and planned integration of PpyRE9h gene. Key: utr1: non-translated region of aprt gene; utr2: 1.4 kb intergenic region from cam operon; and utr3: UTR of dhfr-ts gene; PAC; puromycin resistance cassette. B) PCR confirmation of successful integration of the reporter cassette. Primers 630/637 and 644/645 together confirm the correct integration of the reporter cassette into genome sequences. (see Table 1 for sequences). C) Growth rate of wild-type (black circle) and stably-modified promastigotes PpyRE9h+L.infantum (white circle). Parasites were counted using a Coulter counter. D) In vitro luciferase activity assay of diluted lysates from wild-type (WT) and PpyRE9h+L.infantum promastigotes after PCR confirmation. E) Minimal metacyclic promastigote and infective amastigote number detectable by the IVIS camera. Parasites were loaded in 96-well plates, D-luciferin added and the BLI signal was detected in the IVIS camera. Grey lines indicate detection thresholds determined as the mean (solid line) and mean +2SDs (dashed line) of background luminescence of wells with PBS free-parasites. F) Microscopic image of intracellular PpyRE9h+L.infantum amastigotes infecting PMA-differentiated THP-1 cell line. Bioluminescent amastigotes stained with anti-luciferase IgG (αLuc, red) and Hoechst 33342 DNA stain (H33342, cyan).