Abstract
Cellular control of gene expression is a complex process that is subject to multiple levels of regulation, but ultimately it is the protein produced that determines the biosynthetic state of the cell. One way that a cell can regulate the protein output from each gene is by expressing alternate isoforms with distinct amino acid sequences. These isoforms may exhibit differences in localization and binding interactions that can have profound functional implications. High-throughput liquid-chromatography tandem mass-spectrometry proteomics (LC-MS/MS) relies on enzymatic digestion and has lower coverage and sensitivity than transcriptomic profiling methods such as RNA-seq. Digestion results in predictable fragmentation of a protein, which can limit generation of peptides capable of distinguishing between isoforms. Here we exploit transcript-level expression from RNA-seq to set prior likelihoods and enable protein isoform abundances to be directly estimated from LC-MS/MS, an approach derived from the principal that most genes appear to be expressed as a single dominant isoform in a given cell-type or tissue. Through this deep integration of RNA-seq and LC-MS/MS data from the same sample, we show that a principal isoform can be identified in over 80% of gene products in homogenous HEK293 cell culture and over 70% of proteins detected in complex human brain tissue. We demonstrate that incorporation of translatome data from ribosome profiling further refines this process. Defining isoforms in experiments with matched RNA-seq/translatome and proteomic data increases the functional relevance of such datasets and will further broaden our understanding of multi-level control of gene expression.
Keywords: RNA-seq, Ribosome profiling, Mass-spectrometry, Peptides, Isoforms, Proteogenomics, HEK293, Brain, Expectation Maximisation, Integrative analysis
Introduction
A major challenge in gene expression studies of mammalian systems is the splicing complexity of the transcriptome. Over 90% of human multi-exon protein coding genes can transcribe alternatively spliced mRNAs1; the average gene has the potential to express 3–4 distinct mRNA transcripts, with complex genes potentially generating more than 102. However, several recent observations suggest that the majority of human cell-types and tissues tend to predominantly express a single ‘principal’ RNA transcript3,4. Identification of principal isoforms can yield important biological insights because they dictate the sequence, structure, regulation, and function of the protein(s) produced by the gene. Confident discrimination of these principal isoforms is complicated as each modality of omic data suffers from different biases and confounds with regards to isoform identification. By integrating multiple modes of such data we can attempt to overcome some of these limitations to provide confident principal isoform identification.
Distinguishing between isoforms remains a major challenge for mass spectrometry analysis. Isoforms are most easily distinguished by unique peptide-to-protein identification from peptide spectral matches obtained from LC-MS/MS. Sequence similarity across isoforms limits the number of these unique peptides and corresponding enzymatic cleavage sites within the protein sequence. Current analytical approaches deal with ambiguous or redundant peptide-to-protein matches using a protein grouping feature, which organizes spectral counts into groups representing the entire isoform family5–9. Protein grouping is beneficial because it retains spectral information, but does not provide a solution to isoform ambiguity. Top-down proteomics is a useful strategy for isoform identification, but analysis of intact proteins is often challenging10,11. Targeted mass spectrometry approaches such as parallel and selected reaction monitoring (PRM/SRM) have also been employed to distinguish between isoforms12–18. This method relies on targeting specific masses for peptide identification, and its sensitivity allows identification of low-abundance peptides. While targeted approaches are a promising advancement toward solving the problem of isoform ambiguity, they are still limited by the availability of unique peptides within the protein sequence, and cannot be used on a hypothesis-free basis.
Over the last two decades, genome-wide analysis of nucleic acids has rapidly advanced to the point where we can routinely survey the entire genome, epigenome, and RNA transcriptome of any cellular system. Transcriptome analysis remains the de-facto approach for a genome-wide survey of gene expression while incorporation of proteomic measurements has lagged despite improvements in mass-spectrometry technologies that have put analysis of complete cellular proteomes within reach19–21. Methods for quantifying mRNA-seq at the isoform level have become extremely advanced, despite the challenges inherent in using relatively short reads with non-uniform coverage and a high propensity for multi-mapping. In a 2015 systematic review, 11 of the commonly used tools were found to have comparable consistency and accuracy in isoform quantification, particularly for higher abundance transcripts22. Thus, the most recent improvements to these workflows have focussed on increasing processing speed, decreasing memory usage, and correcting for sequence- and/or position-based biases inherent in RNA-seq experiments.
Recently, studies of the translatome have started to bridge the gap between transcriptome and proteome. Ribosome footprinting measures the dynamic profiles of ribosomes as they are translating mRNA to protein23 and provides additional and often crucial insight into post-transcriptional regulation24–27. Given that protein abundance and modification most closely reflects the biosynthetic state of the cell28,29, it may be advantageous to incorporate translation level data when predicting protein isoforms. Packages for analysis of ribosome footprint data have proliferated in the last two years, but the majority do not explicitly address allocation of footprints to isoforms30–33, instead focusing on improving sensitivity in analysis of translation efficiency. In these approaches, it is common to abstract to the gene-level by choosing a single ‘representative’ transcript; typically, one with either the longest coding sequence or the highest density of footprint reads. Given that calculation of translation efficiency depends on transcript length, mis-identification or naïve aggregation of multi-isoform genes could lead to incorrect quantification. Floor and Doudna used Cufflinks34, a popular package for transcript quantification in an attempt to obtain transcript-level assignments of footprints26, while the Ribomap package uses mRNA transcript abundance to fractionally assign footprints to isoforms35. The Cufflinks approach is inherently limited by the coverage of the footprints and the Ribomap method does not allow for footprints to disagree with mRNA-seq data in the case of significant post-transcriptional regulation.
Here we show that in experiments with carefully matched multi-omic data the vastly greater transcript-resolving power of mRNA-seq can be exploited to enable isoform-level interrogation of the proteome and/or translatome. We adapted a basic expectation maximization technique, now the de-facto standard for mRNA-seq isoform quantification34,36–38, for use with LC-MS/MS and/or ribosome footprint data. By identifying the principal isoform(s) for each gene using the RNA-seq transcriptome quantifications, the EMpire tool (Expectation Maximisation Propagation of Isoform abundance from RNA Expression) set biologically informed priors to guide the assignment of peptides or footprints towards these same isoforms. Use of continuous prior likelihoods represents a continuum of expression that more accurately reflects the underlying biology of a sample than an arbitrary inclusion/exclusion of transcript sequences in a database reference. Divergence from the RNA-seq prediction was allowed if there was sufficient evidence against a particular mRNA isoform due, for example, to substantial post-transcriptional regulation. In concordance with mRNA-seq data3,4, we demonstrated that we can identify a principal isoform in over 80% of gene products in homogenous HEK293 cell culture and over 70% of proteins in complex human brain tissue. Use of these informed priors was beneficial for principal isoform selection in over half of the genes detected, and most, but not all, gene products agreed on the same principal isoform throughout all data modalities.
Experimental Methods
Experimental Design and Statistical Rationale
Each of the HEK293 cell assays was carried out over three biological replicate samples. In all of the HEK cell experimental modalities, a biological replicate consisted of one large 15 cm diameter cell culture dish with HEK cells grown to 75–85% confluence (this equated to approximately 80–100 μL packed cell volume). This sample number was considered appropriate for the exploratory, proof of principle experiments presented here. For the human brain experiment, 5 biological replicates were used; this sample number was based on a previous proteomic study of mouse brain39. No technical replicates were used, as previous experiments showed good consistency across technical replicates.
HEK293 cell culture – Generation of a stable cell line expressing eGFP-L10a
HEK293 cells were transiently transfected with pCMV-EGFP-L10a using Effectene transfection reagent (Qiagen) according to the manufacturer’s protocols. Stably expressing colonies were selected by growth in media containing G418. The pCMV-EGFP-L10a contains the mouse L10a coding sequence, which diverges from the human coding sequence at 71 out of 653 bases, despite ultimately producing the same 100% conserved protein product. This enabled us to assess the ratio of exogenous GFP-L10a to endogenous L10a from our RNA-seq data, which was approximately 1:5 (data not shown) in the cell line (HEK293-L10a) used to produce all data.
Obtaining ribosome-associated RNA (raRNA) by eGFP-L10a immunoprecipitation
raRNAs were obtained according to a modified version of the original bacTRAP immunoprecipitation (IP) protocol40. HEK293-L10a cells were lysed by rotor homogenisation in bacTRAP lysis buffer (20 mM HEPES, 5 mM MgCl2, 150 mM KCl, 0.5 mM dithiothreitol, 100 μg/ml cycloheximide, protease inhibitors, and recombinant RNase inhibitors) plus 2% n-dodecyl-beta-maltoside (n-dodec, Thermo Fisher Scientific). Addition of 2% n-dodec ensures capture of ribosome footprints from both cytosolic and endoplasmic reticulum (ER) associated ribosomes, which in the latter case are otherwise depleted in these preparations41 (and data not shown).
Lysates were cleared by centrifugation at 13,400 × g for 10 min at 4°C, then subjected to IP. For EGFP-L10a IP, 450 μL of BSA blocked MyOne Streptavidin T1 Dynabeads (Thermo Fisher Scientific) were coated with 180 μL biotinylated Protein L, and pre-conjugated to a combination of 75 μg each of the mouse monoclonal antibodies 19F7 and 19C8 (Sloan Kettering Memorial Hospital). Dynabead-antibody complexes were added to the cell lysate and immunoprecipitated overnight. The next day beads were washed 4 times with a high salt wash buffer (10 mM HEPES [pH 7.4], 350 mM KCl, 5 mM MgCl2, 1% NP-40, 0.5 mM dithiothreitol, 100 μg/ml cycloheximide). Bound mRNAs were eluted by resuspending the beads into 700 μL Qiazol and following the manufacturer’s instructions for RNA purification using the miRNeasy kit (Qiagen). Full length total RNA was also prepared by lysing a pellet of HEK293-L10a cells in 700 μL of Qiazol, and using the miRNeasy kit.
Ribosome profiling sample preparation
Ribosome footprints were prepared as described23, with some modifications. Briefly, HEK293-L10a cells were lysed as above in bacTRAP lysis buffer plus 2% n-dodec. Ribosomes were collected by IP with 20 μg of biotin conjugated eGFP monoclonal antibody (Sloan Kettering, as above) complexed with 160 μL Streptavidin Dynabeads. Bead-associated ribosomes were resuspended in 300 μL bacTRAP lysis buffer without RNase inhibitors, and treated with RNaseI as for cell lysates. Digestion was stopped by addition of 10 μL of Superasin (Ambion). Ribosomes were collected by reattachment to the magnet and resuspended in 700 μL Qiazol, and processed as per Qiagen miRNeasy kit instructions. Ribosome footprints were eluted from RNeasy columns in 30 μL RNase free water, then extracted overnight at −80°C following addition of a further 38.5 μL RNase free water, 1.5 μL GlycoBlue (ThermoFisher), 10 μM sodium acetate, and finally 150 μL isopropanol. Footprints were collected by centrifuging at maximum speed on a desktop centrifuge for 30 min at 4°C. Pelleted RNA was air-dried, then run on a 15% TBE Urea Gel (ThermoFisher). A band was cut containing nucleotides of 26–32 nt size. Overnight RNA extraction from the gel pieces, followed by T4 Polynucleotide Kinase (New England Biolabs Inc) treatment of fragments was performed as described23.
RNA-seq library preparation and rRNA depletion
Full length RNA from total cells and raRNA underwent rRNA removal by RiboZero kit (EpiCentre, Illumina), to remove the ~90% of cellular RNA they represent. rRNA depleted RNA was prepared for sequencing according to TruSeq library preparation protocols (Illumina), using random primers to synthesize cDNA. Libraries were run on an Illumina HiSeq 2500 at the Yale Center for Genome Analysis, and paired end 75 nucleotide reads obtained.
Following T4 PNK treatment, ribosome footprints were prepared for sequencing using the NEBNext Small RNA Library Prep kit and the manufacturer’s instructions. This resulted in the use of a single gel extraction step, unlike previous protocols23. After testing various rRNA depletion protocols, we made the decision not to remove rRNA, simplifying the workflow and decreasing the opportunity for investigator introduced variability or end bias.
Mass-spectrometry (MS) proteomics
Frozen pellets of HEK293-L10a cells were lysed by sonication in RIPA buffer plus protease inhibitors. Protein was precipitated from the lysate to remove detergents by chloroform/methanol precipitation. Protein pellets were resuspended in 90 μL of 70% formic acid, and then 360 μL 0.1% TFA was added. Protein was quantified by nanodrop (Thermo Fisher Scientific) and 200μg was aliquoted, dried and reconstituted in in 8 M urea, 0.4 M ammonium bicarbonate, reduced for 30 min at 37°C with 4 mM dithiothreitol, alkylated by incubating for 30 min with 8 mM iodoacetamide, before dilution to 2 M urea and addition of trypsin at a ratio of 1 μg:20 μg total protein. Samples were digested overnight at 37°C, then acidified and desalted on a C18 Macro Spin Column (The Nest Group). Peptides were eluted in 80% acetonitrile/0.1% trifluoroacetic acid (TFA), then dried by Speedvac. The dried pellet was resuspended in Buffer A (10 mM potassium phosphate in 25% acetonitrile solution (pH 3.0)) and separated in the first dimension by Strong Cation Exchange on a 2.1 × 200 mm PolySULFOETHYL A™ column (PolyLC Inc.) via an HP 1090 HPLC Hewlett Packard). Separation was carried out over a linear 118 min gradient with increasing Buffer B (10 mM potassium phosphate, 25% acetonitrile pH 3.0, 1 M potassium chloride) at a flow rate of 200 μL/min. Twenty fractions were collected, pooled into 10 tubes, and each tube desalted using a Ultra-Microspin C18 column (The Nest Group) prior to LC MS/MS. The desalted peptide mixture was reconstituted in Buffer A (Water with 0.1% formic acid) and quantified by Nanodrop (Thermo Fisher Scientific). Peptides were diluted to 0.05 μg/μl, and 5 μL were injection onto the column for each fraction to be analyzed by LCMS/MS. LCMS/MS analysis was performed using an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific) equipped with a Waters NanoACQUITY ultra-performance liquid chromatography (UPLC) system using a Waters Symmetry C18 180 μm by 20 mm trap and a 1.7 μm (75 μm-inner-diameter by 250 mm) NanoACQUITY UPLC column (at 35°C) for peptide separation. Trapping was carried out for 3 min at 5 μl/min in 97% Buffer A (0.1% FA in water) and 3% Buffer B ((0.075% FA in acetonitrile (ACN)) prior to eluting with linear gradients that reached 6% B at 5 min, 35% B at 170 min, and 50% B at 175 min, and 97% B at 180 min for 5 min; then dropped down to 3% B at 186 min for 14 min. Three blanks (1st 100% ACN, 2nd and 3rd Buffer A) followed each injection to ensure against sample carry over.
Mass spectral data were collected over a 300–2000 m/z mass range, with a precursor ion isolation window of 2.0 Da. Data Dependent Acquisition of MS/MS fragmentation (Top 10 with minimum signal of 500 counts) was carried out via High-energy Collisional Dissociation (HCD) with normalized collisional energy of 28 (and activation time of 0.1 sec) and default charge state of 2 for the precursor mass (with charge state rejection of unassigned charge states and 1). Additionally, dynamic exclusion was enabled with repeat count and duration of 1 and 30 seconds respectively. The size of the exclusion list was set at 500 ions for an exclusion duration of 60 seconds. MS1 data were collected in profile mode with 30,000 resolving power setting, while the MS/MS were collected in centroid mode with 15,000 resolving power settings.
RNA-seq read alignment and transcript quantification
Due to our IP of ribosomes, and the decision not to deplete footprint samples of rRNA, we carried out an explicit alignment of the reads to known human rRNA before alignment to the genome.
TotalRNA- and raRNA-seq reads were mapped to the annotated 5S and 45S (chrUn_gl000220) rRNAs using STAR to remove any remaining rRNA contamination. Based on this alignment we observed that residual rRNA could explain on average ~20% of the sequence reads across all totalRNA and raRNA samples. We mapped the remaining ~80% of the RNA-seq reads to the human genome (hg38) and annotated transcriptome (gencode v21) again using the STAR aligner, following roughly the ENCODE alignment parameters (github.com/ENCODE-DCC/long-rna-seq-pipeline/blob/master/DAC/STAR_RSEM.sh). Of the non-default options in STAR, the following are the most important to ensure compatibility with our method:
‘--outSAMtype BAM SortedByCoordinate’ for visualisation in IGV
‘--quantMode TranscriptomeSAM’ for alignments in transcriptome coordinates for eXpress
‘--outFilterMismatchNoverLmax 0.05’ to ensure # mismatches to <5% of the # of mapped bases
Transcript aligned reads were quantified using eXpress.
Ribosome footprint reads were clipped of their 3’ adapter and aligned, like the totalRNA- and raRNA-seq samples above, to the annotated 5S and 45S (chrUn_gl000220) ribosomal RNAs. Removal of rRNA reads was very important to reduce the effect of spurious alignments to the genome. Non-rRNA reads were aligned to the human genome (hg38) and annotated transcriptome (gencode v21) again using the STAR aligner.
Mass-spectrometry spectra alignment
The entire human transcriptome (as defined in gencode21) was in-silico translated, in three frames, to amino acid sequences using the transseq function within the EMBOSS42 software library. Also included in this ‘target’ database were CRAPome43 sequences of likely contaminants (such as Bovine Albumin). For the fractionated HEK cell experiment, MSConvert was used to create a merged.mgf from the proprietary ThermoFisher .raw files from the Orbitrap Elite. The merged.mgf was input to X!Tandem for spectral assignment using default parameters including trypsin cleavage, maximum missed cleavages of 3, minimum ion count of 4, and mapping to the reverse sequence as a decoy.
For the brain experiment, spectra obtained from the Orbitrap Elite, in proprietary ThermoFisher ‘.raw’ files, were processed using MaxQuant44 (v1.5.2.1). Peptides were searched using “trypsin/P” as the digestion enzyme, with a tolerance for up to 2 missed cleavages. This search included a fixed modification, cysteine carbamidomethylation, and two variable modifications, N-terminal acetylation and methionine oxidation. MaxQuant default options were used for mass tolerance; 20 ppm for precursor ions, and 0.5 Da for fragment ions. MaxQuant defaults of FDR corrected p of 0.01 was used for both peptide spectral match and protein identification.
The standard peptides.txt output file from MaxQuant was used as input. Spectra were searched against these transcriptome-derived protein sequences, common contaminant sequences, and a library of reverse ‘decoy’ sequences45.
The distribution of expectation values for spectra with legitimate database hits was compared to the equivalent distribution for spectra assigned to the reverse ‘decoy’ database and a [maximum] expectation threshold was selected for each sample that limited the false discovery rate (FDR) to 1%. I.e. 1% of spectra below this expectation value mapped to the reverse database while 99% mapped to the real database.
Peptides that mapped to more than one distinct genomic locus or to contaminant sequences were discarded from further analysis, however this is not to be confused with peptides assigned to multiple potential isoforms of the same gene which were retained. It is worth noting that many of the peptides identified using the SWISS-Prot reference could be assigned to multiple distinct genes and thus constituted much of the data loss attributed with the use of this reference (data not shown). The transcriptome-derived reference did not suffer as much from multi-mapping, due mainly to less ambiguous gene-isoform relationships.
The analysis described below performs a second round of FDR correction based on decoy mappings and discards multi-locus mapping sequences.
Software implementation and testing
The code for the EMpire expectation-maximization algorithm is freely available and can be found at the Github repo: https://github.com/rkitchen/EMpire
Required inputs to the software are all in common data formats, examples are available in the GitHub repo:
footprinting:
gencode/ENSEMBL genome annotation (.gtf)
footprint read -> transcript alignments (.bam)
[optional] eXpress RNA-seq transcript quants (.xprs)
mass-spec:
gencode/ENSEMBL genome annotation (.gtf)
translated amino-acid sequences for each transcript (.fasta)
spectra -> either transcript X!Tandem output (.xml) or MaxQuant peptides file (peptides.txt)
[optional] spectra MS1 intensities for quantification (.mzXML)
[optional] eXpress RNA-seq transcript quants (.xprs) [OR] footprint EM output (.exprs)
Modifications to an expectation maximization (EM) algorithm
Footprint reads or MS peptides are defined in terms of their binary compatibility to the set of isoforms of a given gene. If a read or peptide has a valid alignment to a transcript it is given a value of 1, or else it is assigned 0. Using this compatibility matrix, I, of all reads/peptides against all transcripts as well as the relative transcript abundances from RNA-seq we can write down the likelihood, P(R1:N|ψ), of observing all 1..N footprint reads, R, given the distribution of the abundances, ψ, of 1..K isoforms:
In the simplest case of the naïve prior, we have no information about which isoform(s) may be responsible for generating the footprint reads and/or peptides so we define the initial distribution of isoform abundances as uniform:
In the case of a non-uniform prior (i.e. from the RNA-seq expression data), ψ is set as the ratios of isoform expressions to the total (cumulative) expression of all transcripts in their parent gene.
Since at the same level of expression isoforms with longer open reading frames will produce more footprint reads (and below saturation longer proteins will produce more identifiable peptides) we also define the probability that the jth isoform will contribute a footprint or peptide based on the length of its coding sequence, l, and its abundance:
For the peptide EM, the effective isoform length is calculated from an in-silico (Trypsin & LysC) digestion (allowing 2 missed cleavages) of the protein to the constituent peptides.
For each gene, we can update the isoform abundances (by Maximum Likelihood Estimation) from the original RNA-seq to new values that best explain the observed footprints. Sampling from these new isoform abundances allows us to assign footprint reads to specific isoforms and so on.
For the ribosome footprints, we further modify the compatibility matrix, I, to reflect the likelihood that a read of this length would be observed with its offset from the coding frame of the transcript. The calculation of this read-position weight matrix is described in the next section. Essentially if 90% of the reads of this read’s length have an observed frame offset of 0.5nt and this read has the same offset to the current transcript then the compatibility is set to 0.9. This allows for the down-weighting of reads that have a spurious frame-offset to the current transcript; in this example if only 2% of all reads of the same length have a frame offset of 1.5nt, which matches that of the current alignment to the current transcript, then the compatibility is set to just 0.02 and this read will have very little positive support for this isoform.
Ribosome footprint frame analysis
Here, we calculated the frame using the offset of the mid-point of the footprint read to the start of the middle-nucleotide of the closest codon triplet. Using this metric and the resulting position-weight-matrix (PWM) of the footprint size vs. codon offset we can infer that the result of incomplete RNase digestion, which will likely differ between footprint preparations, tends to leave additional nucleotides at the 3’ end of the footprint (Figure S4a). We can also use the PWM of read-mids to codon-offsets from single isoform genes to allow the reads to decide for themselves the optimal translation frame for each coding sequence and then ask, as a function of the number of reads mapped to a transcript, what fraction of transcripts are called in the correct frame.
Other bioinformatic & statistical analysis
All RNA-seq statistical analyses, pre-processing, and normalization was performed within the R/Bioconductor scripting environment46. Gene clustering for Figure 4 / Figure S8 was performed using the dynamicTreeCut package, with default parameters except for the minimum cluster sizes which were adjusted for aesthetics. Cluster profiles were computed from the median major isoform fractions of the genes within each cluster. Where referred to explicity in the text, transcript IDs are taken from the October 2014 build of Ensembl: http://oct2014.archive.ensembl.org/index.html.
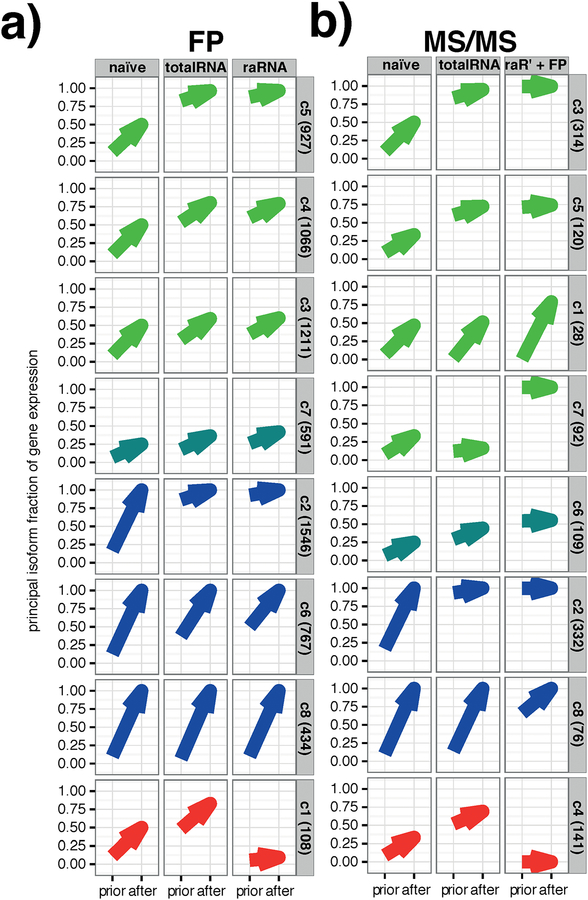
Figure 4 |. A biologically realistic prior improves isoform level interpretation of ribosome footprints and MS/MS peptides.
a) Genes with at least three ribosome footprint (FP) reads clustered into 8 main groups (Fig S8a) based on the result of the EM. The EM algorithm has two components: the naïve or biologically informed prior, and the expectation maximization on the given data. Each plot shows the fraction of total reads assigned to the most dominant isoform (principal isoform fraction) before (prior) and after the EM for each of the 8 groups. The results based on the three available priors are illustrated by the three columns of plots (naive, left; totalRNA, center; raRNA, right). Genes fell into three main groups, those where using a biologically informed RNA-seq prior allowed for selection of a two-fold dominant (y > 0.66) principal isoform (green arrows; clusters 3, 4, 5), those for which the principal isoform was driven entirely by the given footprint data (blue arrows; clusters 2, 6, 8), and those in which the use of a different prior led to different outcomes in terms of the reported principal isoform (“conflicting,” red arrows; cluster 1). An intermediate group also existed (turquoise arrows; cluster 7) for which the informative prior aided in assigning isoforms but was still unable to distinguish between two equally likely principle isoforms. The bracketed number on the right hand side of the plots indicates the number of genes belonging to each cluster.
b) As a) following EM using peptides obtained from mass spectrometry (MS/MS). Peptides clustered into 8 main groups based on the result of the EM (Figure S8b) Here the priors were naive (left), totalRNA (centre), and raRNA+footprints (right); where the latter was the isoform abundance output generated by the ribosome footprint EM using the raRNA prior - the right column in a). Genes fell into three main groups, where using a biologically informed prior allowed for selection of a two-fold dominant principal isoform (green arrows; clusters 1, 3, 5, 7), those for which the principal isoform was driven entirely by the given peptide data (blue arrows; clusters 2, 8), and those in which the principal isoform was conflicted (red arrows; cluster 4). As for a, an intermediate group also exists (turquoise; cluster 6) where the informative prior aided in assigning isoforms but was still unable to distinguish between two equally likely principle isoforms. The bracketed number on the right hand side of the plots indicates the number of genes belonging to each cluster.
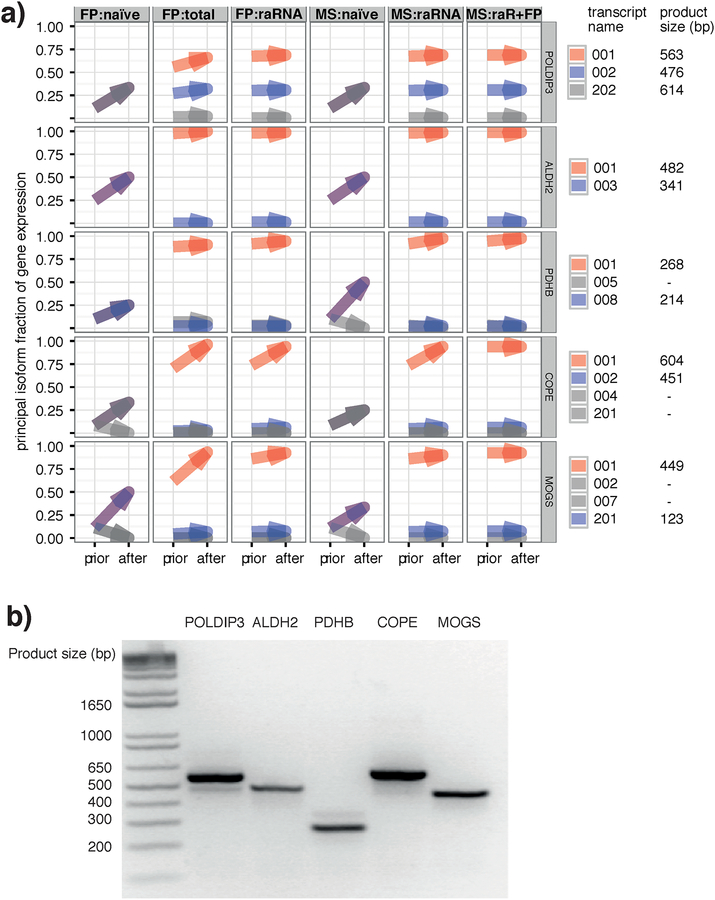
PCR confirmation of principal isoforms from RNA-seq
Total-RNA was extracted from a HEK293-L10a cell pellet as described previously. cDNA was synthesized using SuperScriptIII Reverse Transcriptase (ThermoFisher Scientific) according to Manufacturer’s instructions. PCR primers for selected genes where isoforms were defined by a skipped exon were designed according to the scheme below.
Primers for individual genes were:
| POLDIP3: | AAGTGCAGGATGCCAGAGAG | Fw |
| CAATGGGCTGAGAACAGGCT | Rv | |
| ALDH2: | CCGAGGTCTTCTGCAACCAG | Fw |
| TTGCATCAGGAGCGGGAAAT | Rv | |
| PDHB: | CTGGCTTGGTGCGGAGAC | Fw |
| CCAGCAAAGCCCATCTCTGA | Rv | |
| COPE: | AGAGAGACGTGGAGAGGGAC | Fw |
| CCACTATCCTTGTCTAGCGCC | Rv | |
| MOGS: | CAGGTGTCGCTAACCGGAC | Fw |
| CGGGTCTTCATGCCGAAGTA | Rv |
Standard PCR was performed using Taq DNA polymerase (Invitrogen, Thermo Fisher Scientific). cDNA was diluted 1:10, using 1 μL per 20 μL reaction. 30 PCR cycles were performed, before running the samples on a 1.5% agarose gel.
Data Availability
The mass spectrometry proteomics data and files required to run the analysis described in the paper have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008693.
Results
Integrated experiments for profiling the transcriptome, translatome, and proteome
In order to fully explore the possibilities for isoform-level integration of RNA, ribosome footprint, and proteomic data, we designed a series of assays to be run in parallel on the same cellular sample (Figure 1a). For this proof of principle we used a modified stable human cell-line (HEK293-L10a), collecting RNA-sequencing (RNA-seq) data at two levels, total cellular RNA (‘totalRNA’) as well as immunoprecipitating (IP) only those transcripts engaged by the ribosome (ribosome associated RNA; ‘raRNA’), with the hypothesis that such transcripts may more closely reflect the abundance of protein. We obtained ribosome footprints (‘FP’) through the same (IP)-based approach. Finally, we obtained mass-spectrometry (LC-MS/MS; ‘MS’) proteomic data in ‘discovery’ mode, which relied on fractionating samples to be able to identify more peptides (Supporting Table 1).
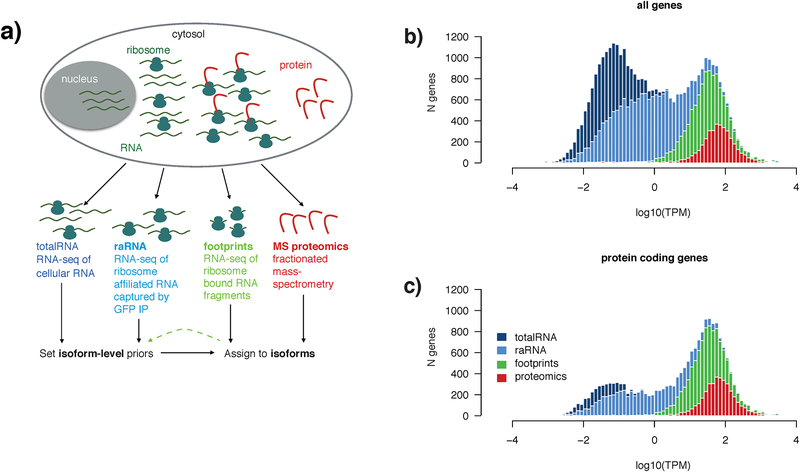
Figure 1 |. Experimental approach to integrated analysis of the transcriptome, translatome, and proteome.
a) Schematic diagram of the experimental approach to multi-modal profiling of the translatome in HEK293-L10a cells. Total-RNA and protein were obtained from lysing whole cells, while ribosome-associated (raRNA) and ribosome footprint RNA were obtained following immunoprecipitation of intact ribosomes from detergent extracted post nuclear supernatant (capturing cytosolic and ER associated ribosomes).
b) RNA-seq of totalRNA (dark blue) captures 60,155 genes (#genes, y-axis) that vary widely in abundance (log10(TPM), x-axis) and biotype. Gene location on the x-axis is defined by the TPM of the total-RNA for that gene, and as such all data shown is a subset of the genes observed by totalRNA. Plotted are genes (note that histograms are overlaid) also observed when profiling raRNA (light blue, 27,977 genes), ribosome footprints (green, lower threshold of 5 ribosome footprints/gene, 11,286 genes), and protein (red, genes with at least 2 peptides, 3833). The bimodal distribution of totalRNA gene expression broadly reflected a distinction between the cohort of mostly low-abundance non-coding RNAs and the higher-expressed protein-coding transcriptome. Genes with TPM > 1 show identical distributions of totalRNA-seq and raRNA-seq gene expression. Genes with at least five ribosome footprints are generally expressed above ~1 transcript per million (TPM). Fractionated LC-MS/MS, where peptides are first separated into pools offline, then analyzed serially, was used to identify proteins.
c) Of the 60,155 total genes captured by total RNA, 19,881 are annotated as protein coding. Ribosome footprints are identified from 56.2% of these protein coding genes, and proteomics samples 18.7%.
These assays differed not only in their molecular target but also their sensitivity. The depth of coverage, in terms of genes detected at all levels of expression, was unsurprisingly by far the greatest in totalRNA (Figure 1b). In our dataset, we observe the vast majority (19,881) of protein-coding genes, of which 11,286 were expressed above 5 transcripts per million (TPM; Figure 1c). raRNA gene expression captured greater than 90% of the protein coding genes detected by total RNA-seq (Figure 1c), while depleting for lower abundance non-coding RNA biotypes, including lncRNAs and processed pseudogenes (Figure S1). We detected at least 5 ribosome footprints from 56.2% of these protein-coding genes, with fractionated ‘discovery’ proteomics identifying 2 or more peptides from 18.7% (Figure 1c).
Intronic reads in totalRNA may act as a confound to transcript quantification by RNA-seq, due to the presence of pre-spliced transcripts from the nucleus (Figure S2a). RNA-seq reads derived from raRNA indeed contained far fewer intronic reads than totalRNA (Figure S2b). We hypothesised that poly-A purification might bring a similar benefit over total RNA, but inspection of data from an ENCODE K562 cell-line (www.encodeproject.org) showed no such reduction in intronic reads (Figure S2c). Notably, the ‘cleaner’ exonic signal from raRNA data led to more consistent transcript quantification across all three biological replicates compared to totalRNA] (Figure S3a,b) This agreement was clearly dependent on both the expression of the gene and on the magnitude of the dominance of the principal isoform (Figure S3c,d). For genes expressed above 5 TPM and with a principal isoform that accounted for more than 50% of the mRNA produced by the gene, principal isoform agreement increased to 93% for totalRNA and to 97% for raRNA (Green lines, Figure S3e,f). While conservative in terms of excluding many non-coding genes, the 5 TPM threshold included 96% of protein coding genes for which we observed footprint reads and/or peptides (Figure 1c).
In the case of ribosome footprints, despite their short fragment size, they contain other useful information that can be leveraged when assigning them to isoforms. The “perfect” cycloheximide frozen ribosome footprint, (consisting of only those nucleotides directly physically protected from the RNase enzyme by the ribosome) is a 28 nucleotide fragment, with the read midpoint situated between nucleotide 1 and 2 of the nearest codon (zero offset, Figure S4a). This consistent fragmentation pattern allows identification of codons from ribosome footprints, and as a result prediction of the open reading frame47 (ORF). Due to variations in the ribosome profiling technique such as incomplete RNase digestion, a range of fragment lengths are obtained. Rather than discard these “imperfect reads,” reads from genes with a single ORF can be used to create a position-weight-matrix (PWM) of read-midpoints to codon-offsets (Figure S4b). The PWM shows the consistency of ORF prediction within each biological replicate, and could be used as a quality control metric to highlight samples with inconsistent RNase digestion. In our hands, consistently digested samples produce a majority of 29 nucleotide length fragments, which correctly predicted the transcript frame 90% of the time (Figure S4b). Incomplete RNase digestion usually resulted in a 3’ base overhang (Figure S4a). Given the consistency highlighted in the PWM, 3 footprints per transcript was sufficient to call the correct frame 75% of the time, rising to greater than 90% accuracy with at least 10 footprint reads (Figure S4c).
Isoform-level integration of RNA-seq, ribosome footprints, and MS peptides
Computational tools for RNA-seq transcript quantification, such as the eXpress algorithm37 used to assess isoform consistency in our data, typically employ an expectation maximization (EM) approach to determine the optimal abundances of each transcript so as to best explain the set of observed sequence reads. It is possible to employ a similar approach to quantifying isoforms based either on LC-MS/MS peptides or on ribosome footprint reads26. Unfortunately, compared to RNA-seq, peptide and ribosome footprint data are much more limited in their capacity to identify specific isoforms due to their smaller size, lower yield, and confinement to the coding sequence (CDS) of the gene. Given that confident isoform discrimination relies largely on peptides or reads that span one or more exon-exon boundaries, identifying the correct isoform from footprints or peptides alone can be problematic. For protein analysis, a random 13 amino-acid peptide (the average observed peptide size in the HEK293 data set) has an average probability of 30% to cross an exon-exon boundary (Figure S5). Despite their increased number, ribosome footprints fare even worse due to their smaller size leading to an average probability of 23% to cross an exon:exon junction. RNA-seq fragments, however, are much longer (especially with paired-end data) and, as such, have a much higher probability (on average 85%) of spanning at least one junction. In principle, it is therefore strongly advantageous to inform an EM model by using totalRNA or raRNA transcript quantifications to set biologically informative priors.
To exploit the increased ability of RNA-seq to quantify the set of expressed transcripts in a sample, we sought to modify a standard EM algorithm to take RNA-seq transcript expression levels (as transcripts per million; TPM) as input to form biologically informative continuous prior likelihoods (priors). These priors represent a continuum of expression that accurately reflects mRNA expression in a sample. These priors were then used to assign short length peptides and/or footprints to isoforms (Figure S6). Unlike a naïve prior (which initially assumes an equal likelihood of any isoform) the RNA-seq derived priors can overcome a large amount of the ambiguity in the set of transcripts/isoforms likely responsible for the observed peptides and/or footprints. In our experiments we applied different combinations of priors depending on the data available. In the HEK293 cell study we assessed both allocation of ribosome footprints using RNA-seq priors (Figure S7), and LC-MS/MS peptides using RNA-seq and footprint isoform likelihood as priors (Figure 2, see methods for more detail on input file formats). This approach also incorporated modifications to the standard EM algorithm to assign more confidence to in-frame ribosome footprint reads, according to the values in the sample-specific PWM (Figure S4).
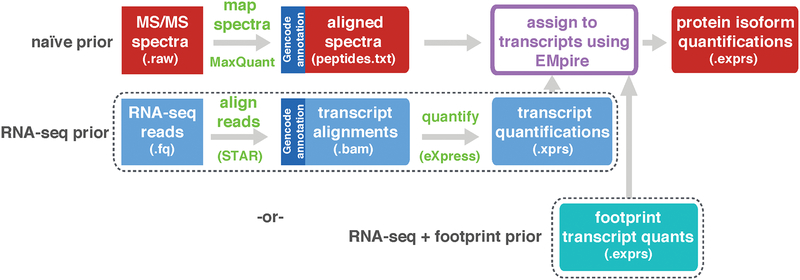
Figure 2 |. Analytical workflow for isoform assignment.
Isoform prediction and assignment for an experiment integrating RNA-seq and proteomics data. Peptide to spectrum matches produced by MaxQuant44 are aligned as transcript coordinates (see Methods if X! Tandem output is a preferred input). The top row is a simple peptide input with no biological prior and the middle is the RNA-seq informed biological prior (transcripts quantified by eXpress37). The bottom row shows integration of RNA-seq, footprint, and MS/MS proteomics, which occurs on a sample-by-sample basis. In this situation the output from the footprint EM (Figure S7) is input to the MS/MS EM.
Using a naïve prior in the HEK293 cell dataset, in 58% of genes neither peptides (blue bars) nor footprints alone (grey bars) could distinguish the principal isoform, instead settling on two or more equally likely isoforms (Figure 3a). In multi-isoform genes for which the naive EM converged on a single principal isoform, this isoform was typically extremely dominant and at least 10-fold more likely than the ‘next-best’ isoform (Figure 3b,c). In the footprint dataset, use of a biologically informative RNA-seq prior established a minimum 2-fold dominant isoform for over 80% of genes. The choice of RNA-seq prior was not critical, as total-RNA and raRNA performed equivalently (Figure 3b). In the LC-MS/MS dataset, use of a biologically relevant raRNA-seq prior resolved a principal isoform for 70% of genes. Inclusion of footprint data in the RNA-seq prior resolved principal isoform ambiguity for a further 8% of proteins compared to raRNA alone (Figure 3c).
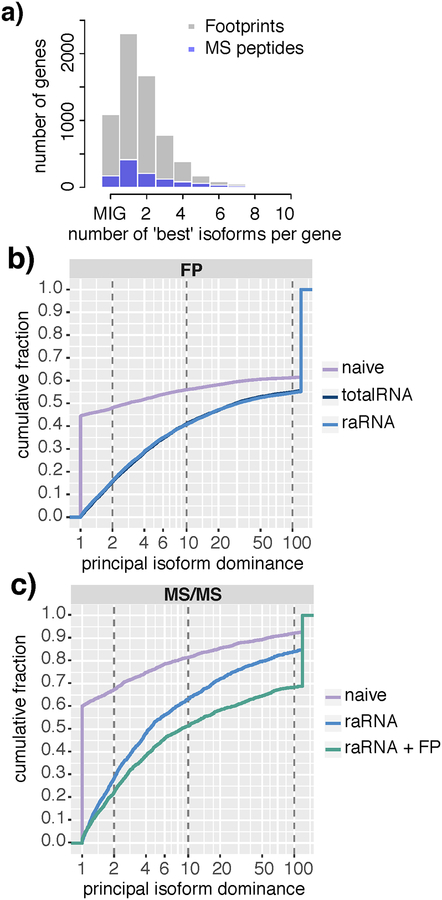
Figure 3 |. Using an RNA-seq prior robustly decreases the ambiguity of footprint and peptide assignment to the principal isoform in multi-isoform genes.
a) Histogram showing the number of equally likely isoforms selected by performing EM with a naïve prior. Genes with only one isoform (“Mono isoform”) are shown as “MIG” on the x-axis. For those multi-isoform genes shown at 1 on the x axis, EM with a naive prior was able to settle on a single dominant isoform. For MS/MS peptide data (blue bars), naive EM was ambiguous as to the likely principal isoform in 58.1% of multi isoform genes. For footprint data (grey bars), 58.2% of multi-isoform genes were ambiguous.
b) Cumulative frequency plot for each of the 9583 genes identified in ribosome footprint data as a result of different biological prior use. The plot shows the fraction of genes with a principal isoform dominance (principal isoform/second isoform) less than X-fold (where x is given on the x axis) For the purpose of clean plotting, principal isoform dominance values greater than 120-fold were capped at 120. A value of 1 reflects a gene in which a single principal isoform cannot be determined. This applies to 45% of genes when a naïve prior is used, versus less than 5% of genes if an RNA prior is used. Vertical dashed lines indicate 2-fold, 10-fold and 100-fold dominant thresholds; for example, 48% of genes have a <2-fold dominant principal isoform using a naïve prior, but this applies to only 16% of genes with an biologically informative prior. As shown by the overlapping lines, use of a total RNA or raRNA prior has an equivalent effect on improving resolution of principal isoform ambiguity, compared to a naïve prior. c) Cumulative frequency plot as per 3b) for each of the 1541 proteins detected by MS/MS. Addition of footprint data to an raRNA prior further improved the resolution of principal isoform ambiguity.
The EM algorithm has two components: the naïve or biologically informed prior, and the expectation maximization on the given data. Using an unsupervised hierarchical clustering and dynamic tree-cut we defined clusters of genes in each of the footprint EM and the proteomic EM that behaved similarly in terms of the ability to resolve a principal isoform on the basis of different priors and/or given data (Figure S8 and Figure 4a–b). These clusters could be generalized into genes for which the biological prior was necessary or beneficial for identification of a two-fold dominant (>66.6 % of gene expression) principal isoform (footprint EM: 57.1% of genes in clusters c3,4,5,7; proteomic EM: 54.7% in clusters c1,3,5,6,7), genes where identification of a dominant principal isoform was driven entirely by the given data (footprint EM: 41.3% of genes in clusters c2,6,8; proteomic EM: 33.7% in clusters c2 and c8), or was conflicting, picking a different principal isoform with different biological priors (footprint EM: 1.6% of genes in cluster c1; proteomic EM: 11.6% in cluster c4).
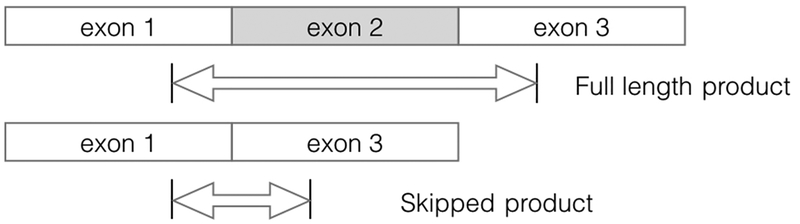
For further validation of the approach, we selected a variety of genes with isoforms containing a single skipped exon, for which the naive prior was unable to help identify a principal isoform but the biological priors appeared to resolve this ambiguity (Figure 5a). We designed PCR primers to amplify the region containing the prospective skipped exon, resulting in products of defined sizes dependent on the presence or absence of the exon. In the example gene highlighted in Figure S9 POLDIP3, use of the RNA-seq prior overwhelmingly suggested the presence of a 2-fold dominant transcript, POLDIP3–001, with a minor transcript of POLDIP3–002, an outcome which was fully consistent with both footprint read locations (Figure S9a), and peptide data (Figure S9a,b). Thus, PCR analysis showed a principal transcript at 563 bp, the product size for transcript POLDIP3–001, with a minor product at 476 bp (transcript POLDIP3–002). For the remaining four cases (ALDH2, PDHB, COPE, MOGS), there was only evidence for the proposed dominant transcript, as predicted by the RNA-seq (Figure 5b).
Figure 5 |. An RNA-seq prior improves isoform assignment in specific genes where a naive prior ties.
a) Detailed illustration of the EM result for 5 selected genes showing differences in the relative isoform abundances of each. In all cases, the biological prior was necessary to resolve the principal isoform (red) and the second isoform (blue) where applicable. To the right, the isoform names are shown along with the expected product size for the PCR validation in b)
b) PCR analysis of the 5 genes selected in panel a) show that, at least at the mRNA level, all agree with the principal isoform inferred by RNA-seq. POLDIP3 (left) also showed evidence for the expression of the second isoform predicted by RNA-seq.
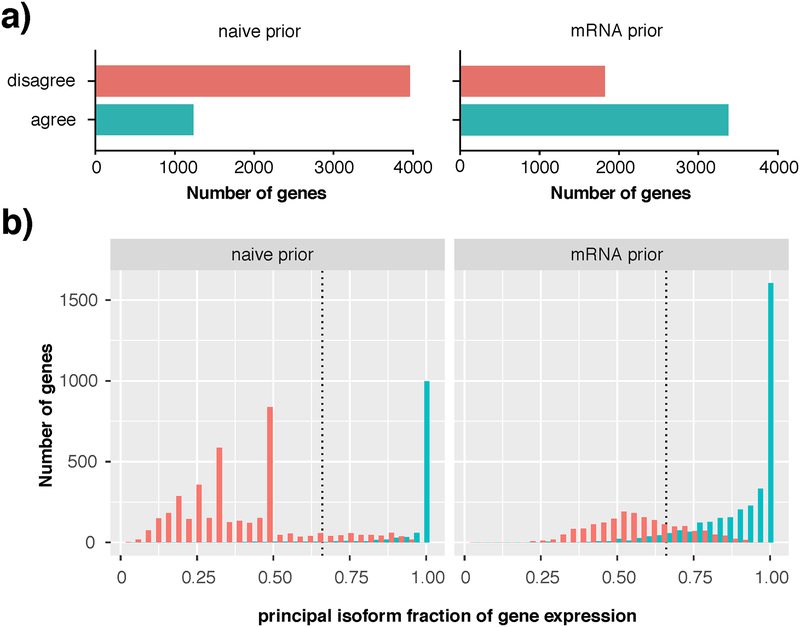
Finally, we showed that this approach was also highly effective when applied to a much more complex dataset with matched mRNA-seq and LC-MS/MS from our recent study of adult human brain regions21. We analyzed 5,197 proteins detected by single-shot LC/MS-MS from the dorsolateral prefrontal cortex (dlPFC) of 5 adult humans, using publicly available mRNA-seq data from the same samples to serve as a biologically informative prior48. In these complex samples, use of the mRNA-seq prior both increased the number of genes where a principal isoform could be consistently defined across all 5 samples (Figure 6a, teal bars), and allowed for selection of a 2-fold dominant principal isoform in an extra 40.6% of proteins (Figure 6b). In 29.8% of proteins the peptide data alone were sufficient for selecting a principal isoform. By using the mRNA prior it was possible to call a 2-fold dominant principal isoform in 70.4% of proteins. Unsurprisingly, as we saw with the HEK293 data, the more dominant a principal isoform was, the more consistently it was called in all five samples (Figure 6b, teal bars).
Figure 6 |. Consistent principal isoform identification in complex human brain samples using a RNA-seq prior.
a) Using an mRNA-seq prior (right) increases consistency of identification of a single minimum 2-fold dominant isoform compared to a naïve prior (left). Red bars (disagree) indicate number of genes where the EM was unable to break a tie between equally likely isoforms, or where different principal isoforms are called in the 5 biological replicates. Teal bars (agree) indicate selection of the same principal isoform in all 5 biological replicates b) Following a naïve prior (left), up to 70% of genes were ambiguous in terms of their principal isoform; red peaks in this dodged histogram are evidence of the algorithm’s failure to break a tie between 2 (x=0.50), 3 (x=0.33) or 4 (x=0.25) equally likely isoforms per gene. Use of the mRNA prior (right) substantially reduced the number of genes with an ambiguous principal isoform, with ~70% of genes reporting a 2-fold or greater dominant isoform (x>0.66). Teal bars indicate genes for which the same principal isoform was called in all five biological samples.
Conclusions
The eukaryotic genome can produce an enormous repertoire of mRNA products. Isoforms are mRNA transcripts that arise from the same gene that may differ in their transcription start site, exon usage, and untranslated regions1. Each of these transcripts may be subject to different regulatory mechanisms, and result in variable structure and function of the final protein product. It is therefore critical to our understanding of gene expression to consider abundance of isoforms, and not simply genes, in high throughput data. The presence of UTRs in mRNA, and the relatively long reads used in paired end mRNA-seq allows for increasingly reliable definition of isoforms in these data. While ribosome footprints may be found in UTRs, peptides are not produced from these regions, and the shorter sequences output from LC-MS/MS or ribosome footprinting are much less likely to cross isoform defining exon boundaries. Here we have demonstrated that use of continuous biologically relevant priors, obtained from matched isoform-level mRNA-seq22 quantification can lead to a marked improvement of the isoform-assignment of ribosome footprints and LC-MS/MS peptides in over 50% of genes.
The EMpire tool takes experiment-level peptide to spectrum matches from widely adopted, freely available proteomics software (we recommend the peptides.txt file from MaxQuant) and uses transcript quantification data from RNA-seq to set biologically informed priors on a sample-by-sample basis, allowing peptides to be assigned to the most likely protein isoforms in each individual sample. This tool may also be used to assign ribosome footprints to isoforms, and ribosome footprints can be input as priors for proteomic experiments. EMpire has workflow advantages over simply using an individualized reference database with which to match proteins and peptides. With an individualized reference, spectra from each sample would have to be matched to their own personalized reference, an approach incompatible with that used in most spectral alignment software, where data from every sample is grouped together, allowing for improved normalization and comparison between samples. Furthermore, thanks to data generated via large projects such as ENCODE49, it has become clear that the deeper a sample transcriptome is sequenced, the more transcripts are detected. It is not clear exactly what the signal:noise threshold is that allows confident definition of the meaningful expression of a transcript, and as such defining on/off characteristics for mRNA expression is challenging and potentially error-prone. The approach presented here defines and exploits continuous prior likelihoods that reflect the underlying biology, which is superior to the arbitrary inclusion/exclusion of transcript sequences in an individualised protein or peptide reference database.
In our proof of concept study using HEK293 cells, almost 60% of genes have no clear principal isoform using a naïve prior. By using a prior derived from mRNA-seq, we can confidently assign a 2-fold dominant isoform to ~80% of genes detected by peptides or ribosome footprints. The addition of footprint data to the mRNA-seq prior further increases the fraction of proteins with a principal isoform at all levels of dominance (2-, 10- and 100-fold). This suggests that in situations where isoform quantification by RNA-seq is noisy, adding more data modalities to isoform level analysis may improve the confidence of isoform selection. For ~55% of these genes, the biological prior is necessary for definition of the principal isoform. For the vast majority of genes detected by ribosome footprints, the different modalities consistently select the same isoform. This is evidence that the assumptions made at the mRNA level that most cell types or even tissues express a principal, dominant isoform of each gene, are valid for studies at the translational and proteomic level. There are some caveats to this latter point which are highlighted by our study, particularly in the proteomic data, where approximately 10% of genes detected by proteomics harbored a disagreement between the RNA-seq and peptide data. This disagreement may be a technical issue related to the coverage of the proteomic data, and the observability of individual peptides. Where only a small number of peptides are detected from a single protein-coding gene, a peptide that disagrees with the RNA prior has significant power to change the outcome. In such cases it is likely that a substantial amount of the disagreement from proteomic data arises from low protein sequence coverage, and will decrease towards the percent disagreement observed with the footprint data (~1.6%) as the sensitivity of LC-MS/MS continues to increase.
We also explored the performance of this approach using proteomic data from a more complex system. Human dlPFC is an extremely complex tissue, with a layered cytoarchitecture and the presence of a large number of individual cell types50,51. We hypothesized that it may be more difficult to define isoforms in this data, as different cell types may contribute different isoforms for the same proteins. However, the use of mRNA-seq as a prior increased the number of genes with a 2-fold dominant isoform from 30% (naïve prior) to 70%. It is likely that a substantial portion of the 30% of genes remaining uncertain can be accounted for by the contributions of isoforms from varying cell types. It is worth noting that the mRNA-seq data used for this study48 were relatively old 50 nucleotide single-end reads. Despite the limitations of these shorter single end reads in defining isoforms, use of this mRNA-seq data still provided a clear benefit for isoform selection from the proteomic data. As we saw in the HEK293 cell experiment, the principal isoform selection was consistent, with the same principal isoform selected in all five biological replicates in 65% of genes.
While the experimental workflow for assignment of peptides to isoforms is therefore extremely flexible with regards to sample type and file input (see Methods), some of the measurements may be unsuitable or more error prone in certain biosample types. Ribosome profiling data for example can only be reliably obtained from fresh, unfixed tissue. Fixing of samples (such as fixed formalin paraffin embedded samples, FFPE) can make tissue more difficult to homogenize and protease digest, and can introduce novel peptides from sites of formalin-induced protein-protein crosslinks52. Such peptides would not be assessed for isoform allocation as they would not be present in standard references used in peptide matching. It is possible that the loss of such peptides may change the outcome of the algorithm, and thus caution should be used when comparing ‘omics data where preservation or preparation may bias one of the profiling modalities. There is also a possibility that proteins that are processed post translationally, such as secreted peptides cleaved by proteases, may disagree strongly with the most likely mRNA transcript, appearing as “conflicting” with regards to agreement between the prior and outcome. As it is currently unclear whether specific transcript isoform abundance is directly related to proteoform abundance on a global scale, it is important to compare RNA read location and peptide level data assigned by this EM approach with the available literature about downstream protein modifications for key experimental targets.
In addition to an improvement in isoform identification, our approach involves mapping all features back to genomic co-ordinates. Currently, it is surprisingly difficult to combine mRNA-seq data with protein data post-hoc. This is partly a result of using non-comparable reference databases in the mRNA-seq versus proteomic data. Mapping back to genomic co-ordinates therefore provides stability for comparisons through multiple versions of reference databases, and makes comparison between different modalities easier. With small adaptations, it will be possible to use this approach for other high throughput data formats such as HITS-CLIP, methylation, and ChIP-seq experiments. This may also prove useful in model systems with less complete reference annotation than human and mouse, using RNA-seq based technologies to define isoforms identified by proteomics. In these systems, using ribosome footprinting may be particularly helpful as the weightings derived from their frame prediction capabilities can be used to define open reading frames. Integration of these data modalities from early in the analysis will increase the functional salience of these data, minimize artifacts arising from poor comparability of reference databases, and enable us to more fully understand the relationship between mRNA, translation and protein.
Supplementary Material
Figure S1. Non-coding RNA biotypes are depleted in raRNAs
Figure S2. RNA-seq of raRNA suffers much less intronic ‘contamination’ than totalRNA
Figure S3. Principal isoform identification is consistent across biological replicates
Figure S4. Ribosome footprints predict the open reading frame
Figure S5. Paired end 200 nt RNA-seq fragments have a large probability of crossing an exon junction
Figure S6. Analytical approach to integrated analysis of the transcriptome, translatome, and proteome
Figure S7. Analytical workflow for isoform assignment
Figure S8. Use of a biologically informative prior improves isoform level interpretation of ribosome footprints and MS/MS peptides
Figure S9. Detailed summary of alignments and EM performance for POLDIP3
Supporting Table 1. Multi-tabbed excel file containing peptide and protein level data from the HEK cell experiments. Corresponding information for the brain experiments can be found in Carlyle et al21.
EMPIRE data.zip. Input and output data from Empire algorithm for the HEK cell experiment
Scheme 1:
Figure showing design of primers for Main Figure 5b. Primers were designed that were placed either side of a skipped isoform, producing products of a different size depending on the mRNA isoform expressed.
Acknowledgements:
This work was supported by NIDA grant R21DA040454 and NIMH grants U01MH103339, MH106934, and MH110926, by the Yale/NIDA Neuroproteomics Centre (DA018343), by a pilot grant from the Center for Molecular and Epigenetic Research of Cell Types Mediating Addictive Behaviors at The Rockefeller University (5P30DA035756), by NIH SIG grants 1S10OD019967–0 & 1S10ODOD018034–01, and by the State of Connecticut, Department of Mental Health & Addiction Services. This work was also supported by NHGRI grant U24HG009446. B.C. was supported by a 2014 NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation. We thank Jean Kanyo for assisting with mass spectrometry data collection. This paper is dedicated to the memory of Edward Voss, who assisted with sample preparation.
Abbreviations
- LC-MS/MS
Liquid chromatography Tandem mass spectrometry
- PRM
Parallel Reaction Monitoring
- IP
Immunoprecipitation
- raRNA
Ribosome-associated mRNA
- FP
Ribosome Footprint
- MS
Mass spectrometry
- EM
Expectation Maximisation
- CDS
Coding Sequence
- TPM
Transcriptions per million
- dlPFC
dorsolateral pre-frontal cortex
- UTR
Untranslated region
References
- (1).Wang ET; Sandberg R; Luo S; Khrebtukova I; Zhang L; Mayr C; Kingsmore SF; Schroth GP; Burge CB Alternative Isoform Regulation in Human Tissue Transcriptomes. Nature 2008, 456, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Katz Y; Wang ET; Airoldi EM; Burge CB Analysis and Design of RNA Sequencing Experiments for Identifying Isoform Regulation. Nat. Methods 2010, 7, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Djebali S; Davis CA; Merkel A; Dobin A; Lassmann T; Mortazavi A; Tanzer A; Lagarde J; Lin W; Schlesinger F; et al. Landscape of Transcription in Human Cells. Nature 2012, 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gonzàlez-Porta M; Frankish A; Rung J; Harrow J; Brazma A Transcriptome Analysis of Human Tissues and Cell Lines Reveals One Dominant Transcript per Gene. Genome Biol. 2013, 14, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Searle BC Scaffold: A Bioinformatic Tool for Validating MS/MS-Based Proteomic Studies. Proteomics 2010, 10, 1265–1269. [DOI] [PubMed] [Google Scholar]
- (6).Nesvizhskii AI; Keller A; Kolker E; Aebersold R A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem 2003, 75, 4646–4658. [DOI] [PubMed] [Google Scholar]
- (7).Zhang J; Xin L; Shan B; Chen W; Xie M; Yuen D; Zhang W; Zhang Z; Lajoie GA; Ma B PEAKS DB: De Novo Sequencing Assisted Database Search for Sensitive and Accurate Peptide Identification. Mol. Cell. Proteomics 2012, 11, M111.010587–M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Old WM; Meyer-Arendt K; Aveline-Wolf L; Pierce KG; Mendoza A; Sevinsky JR; Resing KA; Ahn NG Comparison of Label-Free Methods for Quantifying Human Proteins by Shotgun Proteomics. Mol. Cell. Proteomics 2005, 4, 1487–1502. [DOI] [PubMed] [Google Scholar]
- (9).Wenger CD; Coon JJ A Proteomics Search Algorithm Specifically Designed for High-Resolution Tandem Mass Spectra. J. Proteome Res 2013, 12, 1377–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kelleher Neil L.; Lin Hong Y.; Valaskovic Gary A.; Aaserud David J.; Fridriksson Einar K., and; McLafferty* FW Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. 1999.
- (11).Siuti N; Kelleher NL Decoding Protein Modifications Using Top-down Mass Spectrometry. Nat. Methods 2007, 4, 817–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Simon R; Girod M; Fonbonne C; Salvador A; Clement Y; Lanteri P; Amouyel P; Lambert JC; Lemoine J Total ApoE and ApoE4 Isoform Assays in an Alzheimer’s Disease Case-Control Study by Targeted Mass Spectrometry (n = 669): A Pilot Assay for Methionine-Containing Proteotypic Peptides. Mol. Cell. Proteomics 2012, 11, 1389–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Picard N; Ratanasavanh D; Prémaud A; Le Meur Y; Marquet P Identification of the udp-glucuronosyltransferase isoforms involved in mycophenolic acid phase ii metabolism. Drug Metab. Dispos 2004, 33, 139–146. [DOI] [PubMed] [Google Scholar]
- (14).Lange V; Picotti P; Domon B; Aebersold R Selected Reaction Monitoring for Quantitative Proteomics: A Tutorial. Mol. Syst. Biol 2008, 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Krastins B; Prakash A; Sarracino DA; Nedelkov D; Niederkofler EE; Kiernan UA; Nelson R; Vogelsang MS; Vadali G; Garces A; et al. Rapid Development of Sensitive, High-Throughput, Quantitative and Highly Selective Mass Spectrometric Targeted Immunoassays for Clinically Important Proteins in Human Plasma and Serum. Clin. Biochem 2013, 46, 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Peggion C; Massimino ML; Biancotto G; Angeletti R; Reggiani C; Sorgato MC; Bertoli A; Stella R Absolute Quantification of Myosin Heavy Chain Isoforms by Selected Reaction Monitoring Can Underscore Skeletal Muscle Changes in a Mouse Model of Amyotrophic Lateral Sclerosis. Anal. Bioanal. Chem 2017, 409, 2143–2153. [DOI] [PubMed] [Google Scholar]
- (17).Ghosh S; González-Mariscal I; Egan JM; Moaddel R Targeted Proteomics of Cannabinoid Receptor CB1 and the CB1b Isoform. J. Pharm. Biomed. Anal 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Barthélemy NR; Gabelle A; Hirtz C; Fenaille F; Sergeant N; Schraen-Maschke S; Vialaret J; Buée L; Junot C; Becher F; et al. Differential Mass Spectrometry Profiles of Tau Protein in the Cerebrospinal Fluid of Patients with Alzheimer’s Disease, Progressive Supranuclear Palsy, and Dementia with Lewy Bodies. J. Alzheimer’s Dis 2016, 51, 1033–1043. [DOI] [PubMed] [Google Scholar]
- (19).Mann M; Kulak NA; Nagaraj N; Cox J The Coming Age of Complete, Accurate, and Ubiquitous Proteomes. Mol. Cell 2013, 49, 583–590. [DOI] [PubMed] [Google Scholar]
- (20).Richards AL; Merrill AE; Coon JJ Proteome Sequencing Goes Deep. Curr. Opin. Chem. Biol 2015, 24, 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Carlyle BC; Kitchen RR; Kanyo JE; Voss EZ; Pletikos M; Sousa AMM; Lam TT; Gerstein MB; Sestan N; Nairn AC A Multiregional Proteomic Survey of the Postnatal Human Brain. Nat. Neurosci 2017, 20, 1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Kanitz A; Gypas F; Gruber AJ; Gruber AR; Martin G; Zavolan M Comparative Assessment of Methods for the Computational Inference of Transcript Isoform Abundance from RNA-Seq Data. Genome Biol. 2015, 16, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ingolia NT; Brar GA; Rouskin S; McGeachy AM; Weissman JS The Ribosome Profiling Strategy for Monitoring Translation in Vivo by Deep Sequencing of Ribosome-Protected MRNA Fragments. Nat. Protoc 2012, 7, 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ingolia NT Ribosome Profiling: New Views of Translation, from Single Codons to Genome Scale. Nat. Rev. Genet 2014. [DOI] [PubMed] [Google Scholar]
- (25).Piccirillo CA; Bjur E; Topisirovic I; Sonenberg N; Larsson O Translational Control of Immune Responses: From Transcripts to Translatomes. Nat. Immunol 2014, 15, 503–511. [DOI] [PubMed] [Google Scholar]
- (26).Floor SN; Doudna JA; Amrani N; Ghosh S; Mangus D; Jacobson A; Arava Y; Wang Y; Storey J; Liu C; et al. Tunable Protein Synthesis by Transcript Isoforms in Human Cells. Elife 2016, 5, 1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Ingolia NT; Lareau LF; Weissman JS Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011, 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Kitchen RR; Rozowsky JS; Gerstein MB; Nairn AC Decoding Neuroproteomics: Integrating the Genome, Translatome and Functional Anatomy. Nat. Neurosci 2014, 17, 1491–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Ebrahim A; Brunk E; Tan J; O’Brien EJ; Kim D; Szubin R; Lerman JA; Lechner A; Sastry A; Bordbar A; et al. Multi-Omic Data Integration Enables Discovery of Hidden Biological Regularities. Nat. Commun 2016, 7, 13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Menschaert G; Fenyö D Proteogenomics from a Bioinformatics Angle: A Growing Field. Mass Spectrom. Rev 2015, n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Koch A; Gawron D; Steyaert S; Ndah E; Crappé J; De Keulenaer S; De Meester E; Ma M; Shen B; Gevaert K; et al. A Proteogenomics Approach Integrating Proteomics and Ribosome Profiling Increases the Efficiency of Protein Identification and Enables the Discovery of Alternative Translation Start Sites. Proteomics 2014, 14, 2688–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Xiao Z; Zou Q; Liu Y; Yang X; Cowling VH Genome-Wide Assessment of Differential Translations with Ribosome Profiling Data. Nat. Commun 2016, 7, 11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Zhong Y; Karaletsos T; Drewe P; Sreedharan VT; Kuo D; Singh K; Wendel H-G; Rätsch G RiboDiff: Detecting Changes of MRNA Translation Efficiency from Ribosome Footprints. Bioinformatics 2017, 33, 139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Trapnell C; Roberts A; Goff L; Pertea G; Kim D; Kelley DR; Pimentel H; Salzberg SL; Rinn JL; Pachter L Differential Gene and Transcript Expression Analysis of RNA-Seq Experiments with TopHat and Cufflinks. Nat. Protoc 2012, 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wang H; McManus J; Kingsford C; et al. Isoform-Level Ribosome Occupancy Estimation Guided by Transcript Abundance with Ribomap. Bioinformatics 2016, 32, 1880–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cappé O; Moulines E On-Line Expectation-Maximization Algorithm for Latent Data Models. Journal of the Royal Statistical Society. Series B (Statistical Methodology), 71, 593–613. [Google Scholar]
- (37).Roberts A; Pachter L Streaming Fragment Assignment for Real-Time Analysis of Sequencing Experiments. Nat. Methods 2013, 10, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Patro R; Duggal G; Love MI; Irizarry RA; Kingsford C Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sharma K; Schmitt S; Bergner CG; Tyanova S; Kannaiyan N; Manrique-Hoyos N; Kongi K; Cantuti L; Hanisch U-K; Philips M-A; et al. Cell Type- and Brain Region-Resolved Mouse Brain Proteome. Nat. Neurosci 2015, 18, 1819–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Heiman M; Kulicke R; Fenster RJ; Greengard P; Heintz N Cell Type-Specific MRNA Purification by Translating Ribosome Affinity Purification (TRAP). Nat. Protoc 2014, 9, 1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Reid DW; Nicchitta CV Primary Role for Endoplasmic Reticulum-Bound Ribosomes in Cellular Translation Identified by Ribosome Profiling. J. Biol. Chem 2012, 287, 5518–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rice P; Longden I; Bleasby A EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [DOI] [PubMed] [Google Scholar]
- (43).Mellacheruvu D; Wright Z; Couzens AL; Lambert J-P; St-Denis NA; Li T; Miteva YV; Hauri S; Sardiu ME; Low TY; et al. The CRAPome: A Contaminant Repository for Affinity Purification–Mass Spectrometry Data. Nat. Methods 2013, 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cox J; Mann M MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol 2008, 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- (45).Craig R; Beavis RC TANDEM: Matching Proteins with Tandem Mass Spectra. Bioinformatics 2004, 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- (46).Gentleman RC; Carey VJ; Bates DM; Bolstad B; Dettling M; Dudoit S; Ellis B; Gautier L; Ge Y; Gentry J; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Calviello L; Mukherjee N; Wyler E; Zauber H; Hirsekorn A; Selbach M; Landthaler M; Obermayer B; Ohler U Detecting Actively Translated Open Reading Frames in Ribosome Profiling Data. Nat. Methods 2015, 13, 165–170. [DOI] [PubMed] [Google Scholar]
- (48).Sousa AMM; Zhu Y; Raghanti MA; Kitchen RR; Onorati M; Tebbenkamp ATN; Stutz B; Meyer KA; Li M; Kawasawa YI; et al. Molecular and Cellular Reorganization of Neural Circuits in the Human Lineage. Science 2017, 358, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Harrow J; Frankish A; Gonzalez JM; Tapanari E; Diekhans M; Kokocinski F; Aken BL; Barrell D; Zadissa A; Searle S; et al. GENCODE: The Reference Human Genome Annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Bernard A; Lubbers LS; Tanis KQ; Luo R; Podtelezhnikov AA; Finney EM; McWhorter MME; Serikawa K; Lemon T; Morgan R; et al. Transcriptional Architecture of the Primate Neocortex. Neuron 2012, 73, 1083–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Poulin J-F; Tasic B; Hjerling-Leffler J; Trimarchi JM; Awatramani R Disentangling Neural Cell Diversity Using Single-Cell Transcriptomics. Nat. Neurosci 2016, 19, 1131–1141. [DOI] [PubMed] [Google Scholar]
- (52).Shi Y; Pellarin R; Fridy PC; Fernandez-Martinez J; Thompson MK; Li Y; Wang QJ; Sali A; Rout MP; Chait BT A Strategy for Dissecting the Architectures of Native Macromolecular Assemblies. Nat. Methods 2015, 12, 1135–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Non-coding RNA biotypes are depleted in raRNAs
Figure S2. RNA-seq of raRNA suffers much less intronic ‘contamination’ than totalRNA
Figure S3. Principal isoform identification is consistent across biological replicates
Figure S4. Ribosome footprints predict the open reading frame
Figure S5. Paired end 200 nt RNA-seq fragments have a large probability of crossing an exon junction
Figure S6. Analytical approach to integrated analysis of the transcriptome, translatome, and proteome
Figure S7. Analytical workflow for isoform assignment
Figure S8. Use of a biologically informative prior improves isoform level interpretation of ribosome footprints and MS/MS peptides
Figure S9. Detailed summary of alignments and EM performance for POLDIP3
Supporting Table 1. Multi-tabbed excel file containing peptide and protein level data from the HEK cell experiments. Corresponding information for the brain experiments can be found in Carlyle et al21.
EMPIRE data.zip. Input and output data from Empire algorithm for the HEK cell experiment
Data Availability Statement
The mass spectrometry proteomics data and files required to run the analysis described in the paper have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008693.