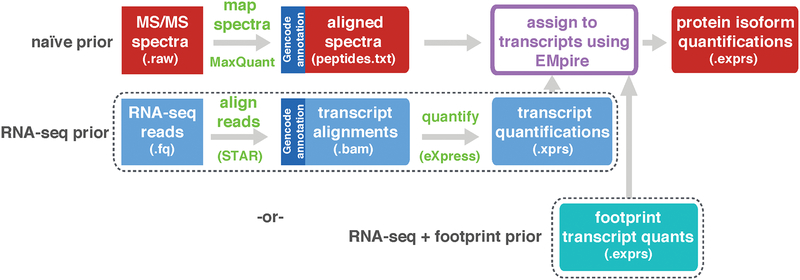
Figure 2 |. Analytical workflow for isoform assignment.
Isoform prediction and assignment for an experiment integrating RNA-seq and proteomics data. Peptide to spectrum matches produced by MaxQuant44 are aligned as transcript coordinates (see Methods if X! Tandem output is a preferred input). The top row is a simple peptide input with no biological prior and the middle is the RNA-seq informed biological prior (transcripts quantified by eXpress37). The bottom row shows integration of RNA-seq, footprint, and MS/MS proteomics, which occurs on a sample-by-sample basis. In this situation the output from the footprint EM (Figure S7) is input to the MS/MS EM.