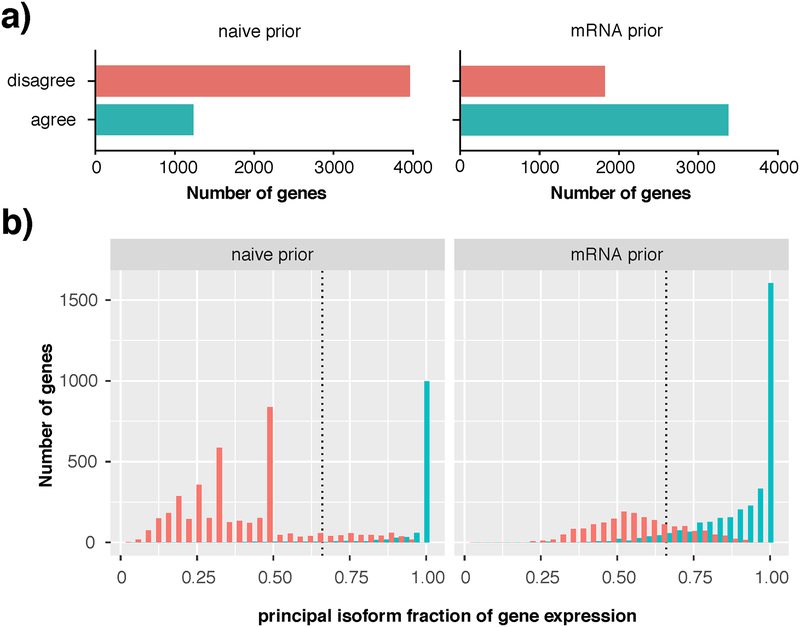
Figure 6 |. Consistent principal isoform identification in complex human brain samples using a RNA-seq prior.
a) Using an mRNA-seq prior (right) increases consistency of identification of a single minimum 2-fold dominant isoform compared to a naïve prior (left). Red bars (disagree) indicate number of genes where the EM was unable to break a tie between equally likely isoforms, or where different principal isoforms are called in the 5 biological replicates. Teal bars (agree) indicate selection of the same principal isoform in all 5 biological replicates b) Following a naïve prior (left), up to 70% of genes were ambiguous in terms of their principal isoform; red peaks in this dodged histogram are evidence of the algorithm’s failure to break a tie between 2 (x=0.50), 3 (x=0.33) or 4 (x=0.25) equally likely isoforms per gene. Use of the mRNA prior (right) substantially reduced the number of genes with an ambiguous principal isoform, with ~70% of genes reporting a 2-fold or greater dominant isoform (x>0.66). Teal bars indicate genes for which the same principal isoform was called in all five biological samples.