Excitatory hippocampal granule cells regulate activation and transition of quiescent neural stem cells during voluntary exercise.
Abstract
The quiescence of radial neural stem cells (rNSCs) in adult brain is regulated by environmental stimuli. However, little is known about how the neurogenic niche couples the external signal to regulate activation and transition of quiescent rNSCs. Here, we reveal that long-term excitation of hippocampal dentate granule cells (GCs) upon voluntary running leads to activation of adult rNSCs in the subgranular zone and thereby generation of newborn neurons. Unexpectedly, the role of these excited GC neurons in NSCs depends on direct GC-rNSC interaction in the local niche, which is through down-regulated ephrin-B3, a GC membrane–bound ligand, and attenuated transcellular EphB2 kinase–dependent signaling in the adjacent rNSCs. Furthermore, constitutively active EphB2 kinase sustains the quiescence of rNSCs during running. These findings thus elucidate the physiological significance of GC excitability on adult rNSCs under external environments and indicate a key-lock switch regulation via cell-cell contact for functional transition of rNSCs.
INTRODUCTION
In the mammalian brain, including rodents and humans, neurogenesis persists throughout adulthood in the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG) and the subventricular zone (SVZ) of the lateral ventricles (1, 2). Neural stem cells (NSCs) reside in these specialized microenvironments, or niches, that maintain them as quiescent, undifferentiated cells to sustain lifelong neurogenesis (3). In the SGZ, adult neurogenesis entails the activation of quiescent type 1 NSCs, also called glia-like radial NSCs (rNSCs) (4). rNSCs divide mostly asymmetrically, producing mitotic, multipotent type 2a NSCs [which are also named amplifying neural progenitors (ANPs)] that are capable of rapid proliferation and differentiation (5). The ANPs are able to differentiate into lineage-committed, proliferative neuronal precursors and, subsequently, neuroblasts as they differentiate into granule cells (GCs) and are integrated into the existing neural network of the hippocampus (6). These newborn neurons derived from the SGZ hereby support memory formation, pattern separation, forgetting, stress buffering, and regulation of affective states (7–10). Deficits in adult hippocampal neurogenesis may contribute to shared facets of many neurodevelopmental, psychiatric, and neurodegenerative disorders (11).
The proliferation, differentiation, and maintenance of rNSCs are orchestrated by a delicate balance of microenvironmental cues. The secreted proteins modulate the balance between quiescent and proliferative NSCs and affect neurogenesis (12, 13). On the other hand, direct cell-cell interaction mediated by integral membrane proteins is critical in stem cell maintenance. For example, direct astrocyte-NSC (14), vascular cell–NSC (15), or mossy cell–NSC (16) interaction controls adult NSC (aNSC) quiescence and regulates hippocampal neurogenesis. An important membrane molecule, Notch, and its ligand can mediate direct interaction between NSCs and neighbor cells and thus play an important role in neurogenesis (17, 18). Noticeably, the microenvironment signals received by rNSCs and the external environmental stimuli (for instance, voluntary running, enriched environment, or fear conditioning) that an organism responds to obviously alter NSC quiescence and neurogenesis (19–22). However, how the extracellular microenvironments precisely transfer the intracellular signals initiated by external environmental stimuli to regulate rNSC properties remains elusive.
The GC neurons serve as prominent components of the SGZ NSC niche and respond to environmental stimuli. The rNSCs with long processes extend from the SGZ toward the molecular layer and ramify there, where they directly interact with these neurons. Previous studies suggest that the quiescence of the rNSCs depends on the tonic release of γ-aminobutyric acid (GABA) from fast-spiking parvalbumin (PV) interneurons in the DG (23), which are activated by GABAergic projection neurons from the medial septum (24). In contrast, strong enhancers of neuronal activity such as systemic injection of kainic acid (KA), a glutamate agonist, increase activation of the rNSCs (25). The neuronal hyperexcitation induced by low and high levels of KA impairs adult hippocampal neurogenesis in the long term (26). These recent findings suggest that NSC quiescence and neurogenesis are controlled directly by the level of activity of the surrounding hippocampal neural network under pathological or artificial stimuli, whereas the functional significance of neuronal activity in the neurogenic niche for rNSC regulation under physiological conditions is poorly understood.
In the present study, we reveal that excitation of glutamatergic GC neurons promotes rNSC activation and neurogenesis in the hippocampal DG region during voluntary running (a physiological training trial). A membrane-bound ligand, ephrin-B3, on GC is identified as a negative regulator for activation of adjacent rNSCs. We further demonstrate that the catalytic kinase activity of the EphB2 receptor in rNSCs is necessary and sufficient for the maintenance of rNSC quiescence during voluntary running. Last, lineage tracing experiments reveal that rNSCs in adult DGs prefer neuronal cell fate upon running attenuated ephrin-B3–EphB2 signal transduction. Our study thus elucidates adjustable intercellular signaling via direct GC-rNSC contact in the neurogenic niche that coordinates glutamatergic neuronal activity and the transition of quiescent NSCs to newborn neurons under physiological conditions.
RESULTS
Excitation of DG neurons and activation of rNSCs during voluntary running
To compare the excitability of DG neurons under different physiological conditions, we treated mice in three training paradigms, including voluntary running, fear stimulation, and enriched environment exposure, which have been previously described to influence neurogenesis (19–22). We performed c-Fos immunofluorescence to indicate neuronal reaction responding to external stimuli among these paradigms and observed activated c-Fos+ cells in a GC layer for each trial. Among them, voluntary running–induced neuronal reactions appeared to be the most noteworthy, reaching a high sustained level after trials spanning 5 to 30 days (fig. S1, A to C, and movie S1). To further examine the running-induced increase in DG granule neuron activity and the temporal relationship between onset of running and granule neuron activity, we introduced in vivo fiber photometry of Ca2+ signals to record the immediate firing of granule neurons during running trials. Adult mice DGs were injected with AAV2/9-hSyn-GCaMP6s followed by optic fiber implantation 6 weeks postnatally (fig. S1, D and E). The data showed that the in vivo Ca2+ signal in granule neurons indicated by fluorescence intensity immediately increased after the onset of running and decreased when the running stopped (fig. S1, F and G, and movie S2), suggesting that the DG cell firing is attributable to the running process.
To examine rNSC behavior and neurogenesis in SGZ in vivo, we identified newborn cells via 5-ethynyl-20-deoxyuridine (EdU) (a thymidine analog) labeling of dividing rNSCs (a combined result of proliferation, survival, and differentiation) in Nestin-GFP transgenic mice (27) over 7 days (once per day) or 30 days (once every 4 days) and carried out immunofluorescence analysis for the activation of rNSCs 1 day after the last injection (Fig. 1A). The activated rNSCs were identified as SGZ cells containing green fluorescent protein (GFP)–positive radial processes with expression of glial fibrillary acid protein (GFAP) and EdU incorporation (Fig. 1B and fig. S2A). We found that voluntary running caused a significant increase of EdU incorporation in rNSCs (Fig. 1, B and C) without affecting GFP-positive rNSC pool size (fig. S2, A to C), suggesting that the long-term physiological running trial induces only mild activation of rNSCs and did not lead to more rapid deletion of rNSCs compared to control mice. Consistently, Ki67 immunostaining showed the same result in the number of activated rNSCs and ANPs (fig. S2, D and E). More nonradial precursor cells lacking GFAP expression (ANPs) were labeled with EdU in the running group and were the progeny of activated rNSCs generated over 7 or 30 days, confirming the effect of running on rNSC transition (Fig. 1C). Furthermore, we observed that the increased level of EdU incorporation in rNSCs in the 7-day running group was restored after 3 weeks of rest following running trials (fig. S2, F to H), indicating that the activation of quiescent NSCs is reversible. We further introduced doublecortin (DCX) to label the neuronal precursors and immature neurons and observed an increase in DCX-positive cells that was accompanied by an increase in EdU-positive cells in the 7-day running group (fig. S3, A and B), suggesting active hippocampal neurogenesis. Two possibilities could account for the increase of neurogenesis: One was the higher proliferation level of rNSCs as shown above, and the other was an increase in the proliferation of intermediate precursor cells (IPCs) themselves. We thus injected EdU after the running trials and euthanized the mice 3 hours later to assess IPC proliferation by identifying colocalization of Tbr2 (an IPC marker) and EdU (fig. S3C). Quantification showed that running mice exhibited a significant increase in Tbr2+ cells, but the proliferation rate between controls and running mice remained unchanged (fig. S3D). These results support the model that running induces rNSC activation in the adult hippocampus and increases neurogenesis.
Fig. 1. Activation of rNSCs for neurogenesis after voluntary running.
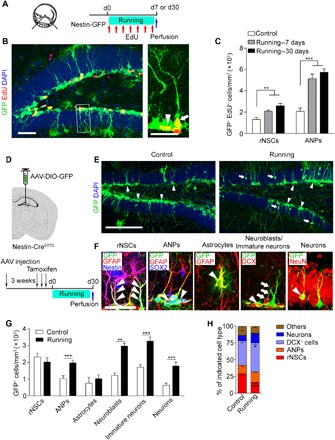
(A) The established protocol used for voluntary running–induced hippocampal rNSC activation and neurogenesis. Ten-week-old mice had access to a running wheel and EdU injection (once per day for 7-day running or once every 4 days for 30-day running) before brain sectioning for immunostaining. d0, day 0. (B) Composite confocal images showing EdU-incorporated rNSCs (arrow) and ANPs (arrowhead). Scale bars, 50 μm (left) and 25 μm (right, zoomed in). DAPI, 4′,6-diamidino-2-phenylindole. (C) Density of EdU-incorporated rNSCs and ANP. In the control, running—7 days, and running—30 days groups, 31, 78, and 87 EdU+ rNSCs and 73, 192, and 208 EdU+ ANP cells of 24, 29, and 27 brain slices were counted, respectively; n = 4 mice for each group. (D) Top: Scheme depicting AAV-DIO-GFP injection into the DG of Nestin-CreERT2 mice. Bottom: Scheme depicting experimental procedure pertaining to injection of viruses into the DG of Nestin-CreERT2 mice. (E) Composite images showing infected GFP+ cells including rNSCs (arrowheads) and young neurons (arrows) in DG regions. Scale bar, 200 μm. (F) Examples of SGZ stem cells and their progeny after infection with AAV, coimmunostained for GFAP (red), Nestin (blue), SOX2 (blue), DCX (red), or NeuN (red). Arrowheads point to processes of infected rNSCs positive for GFAP and Nestin, ANPs positive for SOX2 but negative for GFAP, astrocytes positive for GFAP with astrocyte morphology, neuroblasts positive for DCX with oval morphology, and mature neurons positive for NeuN, respectively. Arrows show infected immature neurons positive for DCX with neuron morphology. (G and H) Graphs show the number/proportion of the different cell types in the niche quantified of all infected cells of Nestin-CreERT2 mice. Control group: 3192 GFP+ cells of 51 brain slices were counted, n = 7 mice. Running group: 5236 GFP+ cells of 53 brain slices were counted, n = 7 mice. Results are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
We next used lineage tracing strategies to explore the effect of running trials on the cell fate of distinct neuronal progenitors in the SGZ. We expressed GFP specifically in the dentate Nestin+ cells by injecting Cre-dependent adeno-associated virus (AAV) vectors (AAV-DIO-GFP) into the DG area in Nestin-CreERT2 mice followed by tamoxifen injections 3 weeks later, which enabled the specific labeling of SGZ rNSCs and the follow-up of their progeny (Fig. 1D). We then evaluated the number of labeled rNSCs (GFAP+/Nestin+ RG-like morphology), ANPs (GFAP−/SOX2+), neuroblasts (DCX+, with oval morphology), immature neurons (DCX+, with neuron morphology), neurons (NeuN+), and astrocytes (GFAP+, with astrocyte morphology) within the GFP+ population in 30-day running mice and observed an increase in the number of ANPs, neuroblasts, immature neurons, and neurons except for rNSCs and astrocytes (Fig. 1, E to G). Quantitation of the proportion of this population also showed increased DCX+ cells and neurons among GFP+ cells (Fig. 1H), indicating that running trials induce a transition toward neuronal fate.
Excited dentate GCs regulate rNSC property during voluntary running
We next addressed which neuronal subpopulation in DG was responsible for voluntary running. We checked c-Fos signals in different subtypes of neurons following the running trial and found that voluntary running mainly activated glutamatergic neurons rather than GABAergic neurons in the niche (fig. S4, A to D). To further assess the functional impact of these glutamatergic neurons on rNSCs in vivo, we injected Cre-dependent designer receptor exclusively activated by designer drug (DREADD) AAV-DIO-hM3Dq-mCherry or AAV-DIO-mCherry, as a control, into the DG of adult CaMKII-Cre mice, which allowed us to precisely activate these hM3Dq-expressed neurons by applying the specific ligand clozapine-N-oxide (CNO) (Fig. 2A). We validated the injection in the DG area (Fig. 2B) and observed that these mCherry-labeled cells were vGluT2 neurons rather than Nestin-GFP, DCX, and GAD65/67 cells (fig. S5, A and B). The small number of Nestin-GFP and DCX cells infected by AAV (~0.5% for Nestin-GFP, 0.2% for DCX cells) indicates that the mistarget of AAV in Nestin-GFP or DCX cells was rare and would not affect transition of rNSCs. We also detected the excitation of GAD65/67 cells with c-Fos staining, and no difference between the control group and the CNO group was observed in these neurons (fig. S5C). Further immunostaining with somatostain (SST) and PV markers in the AAV-injected brain revealed that mCherry+ PV cells were undetectable, and few mCherry+ SST cells were observed (fig. S5D). This result suggested that the number of GAD65/67+ cells possibly infected by the AAV was negligible with respect to the neuronal circuit. To activate these neurons in an approximately physiological level, we tested the effect of different doses of CNO (0.05 to 0.8 mg/kg intraperitoneally) and found that following 0.2 or 0.4 mg/kg CNO administration, the DG GC layers exhibited levels of c-Fos+ cells similar to those of voluntary running (fig. S6). We then used the doses of CNO to activate these neurons and labeled the proliferating cells with EdU and observed a marked increase in the number of EdU-incorporated rNSCs in either the 0.2 or the 0.4 mg/kg CNO group (Fig. 2C), while in the control virus–injected groups, neither the number of EdU+ rNSCs nor that of Tbr2+ cells/DCX+ cells was obviously affected by CNO administration (fig. S7, A to C). Moreover, the total number of DCX+ cells was also significantly up-regulated in a dose-dependent manner with the increase in EdU+ cells after CNO treatment (fig. S7, D and E). Furthermore, we assessed IPC proliferation by identifying colocalization of Tbr2 and EdU and found that although CNO treatment activated rNSCs to generate a large number of Tbr2+ cells, the proliferation rate of IPCs itself was not altered similar to that in running mice (fig. S7E). These results indicate that excited glutamatergic GCs activate rNSCs and promote neurogenesis.
Fig. 2. Dentate GCs excited by chemogenetic approach promotes rNSC activation.
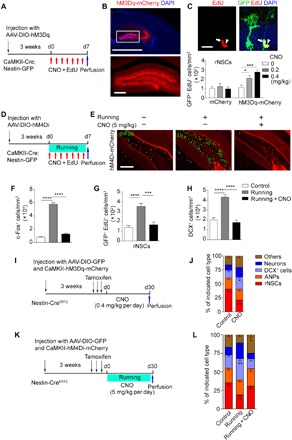
(A) Experimental paradigm for in vivo chemogenetic stimulation. (B) Composite images showing infected neurons in DG regions. Scale bars, 2000 μm (top) and 200 μm (bottom). (C) Composite confocal images showing EdU-incorporated rNSCs (arrows) and ANPs (arrowheads). Scale bar, 25 μm. Density of EdU-incorporated rNSCs in the DG after CNO treatment at concentrations of 0, 0.2 and 0.4 mg/kg. mCherry: 77, 86, and 79 EdU+ rNSCs of 56, 54, and 56 brain slices in the 0, 0.2, and 0.4 mg/kg group were counted, respectively; n = 7 mice for each group; hM3Dq-mCherry: 88, 217, and 256 EdU+ rNSCs of 49, 53, and 56 brain slices in the 0, 0.2, and 0.4 mg/kg group were counted, respectively; n = 7, 8, and 8 mice (bottom). (D) Experimental paradigm for in vivo chemogenetic inhibition by AAV-DIO-hM4Di-mCherry virus injection in CaMKII-Cre; Nestin-GFP mice under voluntary running. (E) Images of c-Fos+ cells in the DG following chemogenetic inhibition under voluntary running trials. Scale bar, 200 μm. (F to H) Quantification of c-Fos+ cells and EdU-incorporated rNSCs and DCX+ cells in the DG of the control and running group following chemogenetic inhibition under voluntary running trials. Control, Running, and Running + CNO groups: 1488, 8574, and 1083 c-Fos+ cells; 87, 225, and 97 EdU+ rNSCs; and 3597, 9086, and 4023 DCX+ cells of 49, 55, and 58 brain slices were counted, respectively; n = 7, 8, and 8 mice. (I) Scheme depicting the experimental procedure for 30-day lineage tracing under GC excitation. (J) The graph shows the proportion of the different cell types in the niche quantified of all GFP+ cells of Nestin-CreERT2 mice. Control group: 4021 GFP+ cells of 51 brain slices were counted; n = 9 mice; CNO group: 5781 GFP+ cells of 49 brain slices were counted; n = 8 mice. (K) Scheme depicting the experimental procedure for 30-day lineage tracing in running mice under GC inhibition. (L) The graph shows the proportion of the different cell types in the niche quantified of all GFP+ cells of Nestin-CreERT2 mice. Control group: 2877 GFP+ cells of 44 brain slices were counted; n = 7 mice; Running group: 4016 GFP+ cells of 45 brain slices were counted; n = 7 mice; Running + CNO group: 2511 GFP+ cells of 48 brain slices were counted; n = 6 mice. Results are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To investigate whether the excitation of glutamatergic GCs accounts for the activation of rNSCs and neurogenesis in voluntary running, we labeled the dentate GCs with Cre-dependent AAV-DIO-hM4Di-mCherry (Fig. 2D and fig. S7F), which allows the selective inhibition of neuronal activity by CNO injection following running trials. We validated neuron activity by c-Fos immunofluorescence and found that the c-Fos+ neurons excited by voluntary running were almost eliminated after CNO injection (Fig. 2, E and F). We thus examined NSC quiescence and neurogenesis upon the simultaneous inhibition of these neurons in the running mice and observed restored levels of rNSC activation and DCX+ cells compared to the control group (Fig. 2, G and H). We also injected the mice with the control virus followed by CNO (5 mg/kg) treatment and did not see changes in the number of EdU+/activated rNSCs in either control or running mice (fig. S7, G and H). The CNO injection did not affect running wheel activity exhibited by the average daily running distance (fig. S7I). These data suggest that inhibition of dentate GCs reverses the activation of rNSCs in running mice.
To further decipher the cell fate of distinct neuronal progenitors upon excitation of GCs, we labeled the dentate Nestin+ cells by using a linage tracing strategy and specifically stimulated glutamatergic neurons by injecting them with AAV-CaMKII-hM3Dq-mCherry followed by CNO treatment for 30 days (Fig. 2I). Quantitation of the proportion of subpopulation among GFP+ cells showed increased DCX+ cells and neurons and decreased rNSCs (Fig. 2J), indicating that a neuronal fate shift occurs upon activation of glutamatergic neurons. Moreover, we examined a lineage shift of rNSC progeny upon the simultaneous inhibition of these neurons with AAV-CaMKII-hM4Di-mCherry in voluntary running mice and observed a restoration in proportion of either DCX+ cells or neurons among GFP+ cells (Fig. 2, K and L). These data suggest that excitation of dentate GCs upon voluntary running promotes fate transition of rNSCs toward neurons.
Ephrin-B3 is identified as a regulator in DG upon running-induced neuron excitation
The leading role of activation of surrounding excitatory GCs in rNSCs led us to ask whether the direct GC-rNSC interactions were involved in the local niche. We simulated three-dimensional (3D) reconstruction of consecutive confocal Z stack images in Nestin-GFP mice that had been injected with AAV-CaMKII-mCherry in the DG. The rNSCs were labeled green and the GCs were labeled red. The 3D image and movie showed a direct contact of rNSCs and GCs in DG areas (Fig. 3A and movie S3). We analyzed previous sequencing results (28), verifying that gene expression changed in the DG after 30 days of voluntary running. We found that, after the running treatment, these changed genes were significantly enriched in Gene Ontology (GO) terms such as cell adhesion, receptor binding, and membrane, suggesting that certain membrane molecules in excitatory neurons may play important roles in voluntary running (Fig. 3B). Therefore, we focused on these associated genes and observed 255 common genes by comparing the three GO terms. These genes were further compared with 1802 differentially expressed genes (DEGs) in the GC layer of DG after running to obtain 46 overlapping genes, which involves 9 genes encoding cell membrane proteins expressed in the nervous system (Fig. 3C and fig. S8).
Fig. 3. Ephrin-B3 is down-regulated in the excited DG region under voluntary running.
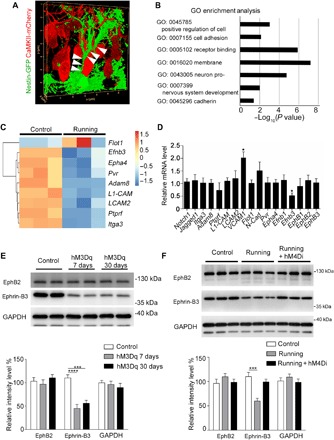
(A) 3D reconstruction of confocal images of rNSCs and GCs in Nestin-GFP mice that were injected with AAV-CaMKII-mCherry. The confocal Z stack images were simulated to 3D images with LAS X software. The arrowheads indicate the direct contact areas of rNSCs and GCs. (B) Results of GO Enrichment analysis on the set of 1802 DEGs. The length of each bar indicates the log10 (P value), and the vertical axis shows significantly enriched terms. (C) The selected nine genes with differential expression are shown in the heat map. (D) mRNA from DG excited by the chemogenetic approach was used to detect differential expression of genes selected from previous sequencing data after running (28). (E) The expression of ephrin-B3 in DG tissue from control and hM3Dq mice was detected by Western blot. (F) The expression of ephrin-B3 in DG tissue from control and hM4Di mice under voluntary running treatment was detected by Western blot. Results are presented as means ± SEM. *P < 0.05; ***P < 0.001; ****P < 0.0001.
We further validated the associated membrane genes, including the nine genes from sequencing analysis and eight additional neuronal genes that have been reported (29), by using a chemogenetic approach in GCs (Fig. 3D). We found that vascular cell adhesion molecule 1 (VCAM1) and ephrin-B3 were obviously altered among these genes (Fig. 3D), and ephrin-B3 is the only one that is highly expressed in the GC layer, as shown in the Allen Brain Atlas database and in previous studies (30, 31). Consistently, we also saw a down-regulated ephrin-B3 level in the DG by Western blot after treatment of either hM3Dq chemogenetic stimuli or voluntary running (Fig. 3, E and F). Furthermore, inhibition of dentate GCs by hM4Di stimulation rescued ephrin-B3 expression to the control level in the DG in voluntary running mice (Fig. 3F). These data indicate that the expression of ephrin-B3 is responsive to voluntary running, which gives rise to the presumption that the regulation of NSC property is likely initiated though GC-rNSC contact.
Ephrin-B3 serves as a GC membrane–bound ligand for rNSC regulation
Ephrin-B–EphB signaling is traditionally known as a membrane-bound ligand-receptor system that controls the spatial organization of cells and their projections by modulating intercellular attractive and repulsive forces (32). In particular, ephrin-Bs and their EphB receptor tyrosine kinases mediate cell-cell bidirectional communication between neighboring cells to control cell migration, survival, and proliferation through multiple effector pathways (33). To examine the role of ephrin-B3 in regulating rNSCs in vivo, we used Efnb3−/− mice and treated them with EdU over 7 days for NSC immunostaining and counting. We observed more activated rNSCs and ANPs in Efnb3−/− mice by EdU incorporation in NSCs (Fig. 4, A to C). Moreover, these Efnb3−/− mice showed higher total numbers of both EdU+ cells and DCX+ cells than wild-type (WT) mice, revealing brisk neurogenesis in the absence of ephrin-B3 (fig. S9, A and B). As a binding ligand of the EphB receptor, ephrin-B3 is capable of binding EphB receptors to stimulate forward signaling and serves as a receptor possibly to transduce reverse signals into the cells on which it is expressed (31, 34). To distinguish the two directions of signaling, we examined the neurogenesis in Efnb3LacZ/LacZ mice, an ephrin-B3 mutant expressing a truncated ephrin-B3–β-galactosidase (β-gal) fusion protein with the same temporal, spatial, and subcellular pattern as the endogenous WT protein. The mutant retains the extracellular and transmembrane domains to provide ligand-like activity but is unable to transduce reverse intercellular signals. Unlike the Efnb3−/− mice, Efnb3LacZ/LacZ mice showed no difference compared to WT mice (fig. S9, A and B), suggesting that ephrin-B3 serves as an external ligand in the niche to stimulate EphB-mediated forward signaling into rNSCs.
Fig. 4. Ephrin-B3 is involved in the shift from quiescent rNSCs to activated NSCs.
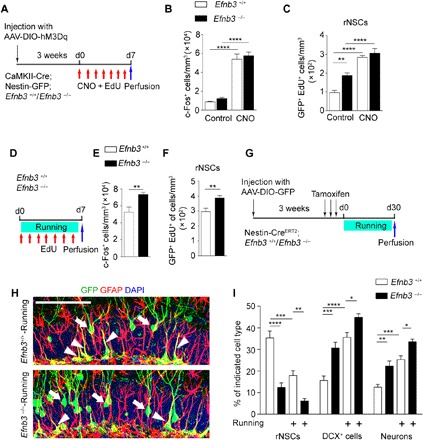
(A) Experimental paradigm for chemogenetic stimulation by AAV-DIO-hM3Dq-mCherry injection in the DG of CaMKII-Cre; Nestin-GFP; Efnb3−/−mice. (B and C) Quantification of the number of c-Fos+ and EdU-incorporated rNSCs after hM3Dq stimulation. In the Efnb3+/+, Efnb3−/−, Efnb3+/+ + CNO, and Efnb3−/− + CNO groups, 1325, 2314, 6903, and 6721 c-Fos+ cells and 65, 176, 189, and 203 EdU+ rNSCs of 43, 39, 44, and 49 brain slices were counted, respectively; n = 6, 6, 7, and 6 mice. (D) Experimental paradigm for the roles of ephrin-B3 in rNSC activation under voluntary running. (E and F) Quantification of the number of c-Fos+ and EdU-incorporated rNSCs in Efnb3 +/+ and Efnb3−/− mice after voluntary running. In the Efnb3+/+ and Efnb3−/− running groups, 4627 and 6019 c-Fos+ cells and 126 and 167 EdU+ rNSCs of 30 and 31brain slices were counted, respectively; n = 4 mice for each group. (G) Scheme depicting the experimental procedure for lineage tracing in running Efnb3−/− mice. (H) Composite images showing infected GFP+ cells in the DG regions of Efnb3+/+-Running and Efnb3−/−-Running mice. Scale bar, 100 μm. (I) The graph shows the proportion of the different cell types in the niche quantified of all GFP+ cells of Nestin-CreERT2 mice. Control group: 3024 GFP+ cells of 38 brain slices were counted; n = 5 mice; Efnb3−/− group: 3689 GFP+ cells of 43 brain slices were counted; n = 7 mice; Efnb3+/+-Running group: 5019 GFP+ cells of 55 brain slices were counted; n = 8 mice; Efnb3−/−-Running group: 4716 GFP+ cells of 49 brain slices were counted; n = 8 mice. Results are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To investigate whether the alteration in rNSC behavior upon GC excitation is via down-regulation of neuronal ephrin-B3, we injected AAV-DIO-hM3Dq-mCherry into WT or Efnb3−/−; CaMKII-Cre; Nestin-GFP mice and treated these mice with CNO for 7 days (Fig. 4A). We observed a comparable increased number of c-Fos–labeled GCs and activated rNSCs, up to two- to threefold of the WT control level, in both WT and Efnb3−/−mice upon CNO administration, suggesting that ephrin-B3 plays a major role in preventing rNSC activation upon GC excitation in the adult hippocampus (Fig. 4, B and C). Consistent with neuronal excitation via the chemogenetic approach, we performed voluntary running trials and saw a significant increase in the number of c-Fos+ GCs in Efnb3−/− mice as compared with WT mice (Fig. 4, D and E), and this was accompanied by a greater increase in EdU-incorporated rNSCs in Efnb3−/− mice (Fig. 4, D and F). These results indicate that ephrin-B3 functions in coordination with GC excitation and rNSC activation during voluntary running.
We also carried out lineage tracing of adult rNSCs in Nestin-CreERT2 mice that were crossed with Efnb3−/− mice to examine the effect of ephrin-B3 on the progeny of stem cells after voluntary running (Fig. 4, G to I). We observed decreased rNSCs and increased DCX+ cells/neurons among GFP+ cells in Efnb3−/− mice (Fig. 4, H and I), but not in Efnb3LacZ/LacZ mice (fig. S9, C to E), indicating that ephrin-B3 serves as an external ligand in GCs to promote adult neurogenesis. Furthermore, after the 30-day running trial, an increase in DCX+ cells and neurons among GFP+ cells but a decrease in rNSCs were observed in both lineage-traced WT or Efnb3−/− mice (Fig. 4I). Together, the data suggest that ephrin-B3 on GC membranes serves as a negative regulator in the niche, affecting quiescent NSCs and preventing transition of rNSCs to neurons.
The kinase activity–dependent EphB2 pathway in rNSCs is involved
To determine whether EphB receptors mediated the intercellular regulation of GC on adult rNSCs, we examined expression of EphB receptors in rNSCs of hippocampal SGZ using EphB2LacZ knock-in mouse line (35) that contains membrane-localized C-terminal truncated EphB2–β-gal fusion protein as a reporter for EphB2 expression. Our analysis revealed that EphB2 was strongly expressed in Nestin-GFP+ rNSCs that were identified with a SOX2 marker (fig. S10A). We further clarified that EphB2 in radial processes of rNSCs was colocalized with GC ephrin-B3 visualized by β-gal immunostaining in Efnb3LacZ/+ mice (fig. S10B). These results indicate that EphB2 is an rNSC-expressing receptor that directly contacts GC ephrin-B3 in adult hippocampus.
To test whether EphB2 intracellular signals play a role in the regulation of adult rNSCs, we used EphB2 null and function domain truncated mutants (35) and examined the proliferation and differentiation of rNSCs. We observed an increased number of EdU-incorporated rNSCs (Fig. 5, A and B) and DCX+ cells (Fig. 5, A to C) in SGZ of adult EphB2−/− mice, suggesting that EphB2 is required for the maintenance of quiescent rNSCs and regulation of neurogenesis. Since the EphB receptor is also capable of transducing bidirectional signals, EphB2 mutant mice with various cytoplasmic domain mutations were examined to further clarify whether the EphB2-mediated forward signaling is involved in regulating the number of rNSCs. These mice express a truncated EphB–β-gal fusion protein as a substitute for the majority of their respective cytoplasmic segment, as mentioned above, or an intracellular segment carrying mutations disrupting EphB2 kinase activity (K661R) or its PDZ domain–binding motif (ΔVEV) (36) to disrupt forward signaling but retain the extracellular and transmembrane domains to provide ligand-like activity to transduce reverse signaling. The immunostaining results revealed that EphB2LacZ/LacZ and EphB2K661R/K661R mutant mice but not EphB2ΔVEV/ΔVEV mutant mice showed an increased number of EdU-incorporated rNSCs and DCX+ cells similar to EphB2−/− mice (Fig. 5, A to C). We further assessed IPC proliferation by identifying colocalization of Tbr2 and EdU and found that disrupting EphB2 kinase activity in rNSCs led to NSC activation but did not affect the proliferation rate of IPC (fig. S10, C to E). These data indicate that kinase activity–dependent EphB2 forward signaling is essential to sustain NSC quiescence.
Fig. 5. Active EphB2 kinase is required and sufficient for the maintenance of quiescent rNSCs.
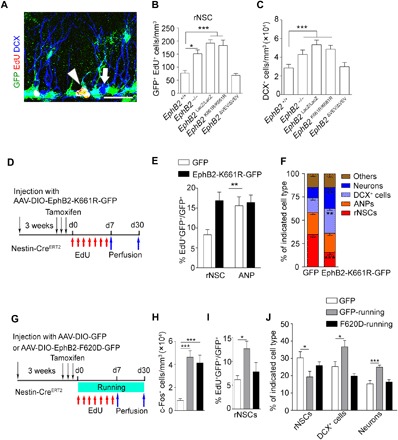
(A) Composite confocal images showing EdU-incorporated rNSCs and DCX+ cells. Scale bar, 25 μm. (B and C) Quantitative analysis showing that EphB2−/−, EphB2LacZ/LacZ, and EphB2K661R/K661R mice have increased numbers in EdU-incorporated rNSCs and DCX+ cells compared with control mice, but the EphB2ΔVEV/ΔVEV remain unchanged. (n = 3 for each group). (D) Scheme depicting the experimental procedure for rNSC activation and lineage tracing in mice after injection with AAV-DIO-EphB2-K661R-GFP in the DG region. (E) The graph shows the proportion of EdU+ rNSCs or EdU+ ANP cells among virus-infected cells of Nestin-CreERT2 mice injected with AAV-DIO-EphB2-K661R-GFP. In the GFP and EphB2-K661R-GFP groups, 35 and 78 EdU+GFP+ rNSCs and 102 and 143 EdU+GFP+ ANPs of 55 and 56 brain slices were counted, respectively; n = 8 mice for each groups. (F) The graph shows the proportion of the different cell types in the niche quantified of all GFP+ cells of Nestin-CreERT2 mice. In the GFP and EphB2-K661R-GFP groups, 3147 and 4468 GFP+ cells of 49 and 51 brain slices were counted, respectively; n = 7 and 8 mice. (G) Scheme depicting the AAV-DIO-EphB2-F620D-GFP injection into the DG of the Nestin-CreERT2 mouse line under voluntary running for 7 or 30 days. (H) Quantification of the density of c-Fos+ cells in Nestin-CreERT2 mice injected with GFP or EphB2-F620D-GFP virus after 7-day voluntary running. In the GFP, GFP-running, and F620D-running groups, 1204, 4283, and 4768 c-Fos+ cells of 51, 50, and 55 brain slices were counted, respectively; n = 7, 8, and 7 mice. (I) The graph shows the proportion of EdU+ cells of GFP+ rNSCs of Nestin-CreERT2 mice injected with AAV-DIO-GFP or AAV-DIO-EphB2-F620D-GFP after 7 days of running. In the GFP, GFP-running, and F620D-running groups, 69, 153, and 129 EdU+GFP+ rNSCs of 51, 50, and 55 brain slices were counted, respectively; n = 7, 8, and 7 mice. (J) The graph shows the proportion of rNSCs, DCX+ cells, and NeuN+ cells of all GFP+ cells of Nestin-CreERT2 mice injected with AAV-DIO-GFP or AAV-DIO-EphB2-F620D-GFP after 30 days of running. In the GFP, GFP-running, and F620D-running groups, 1796, 2879, and 1903 GFP+ cells of 41, 41, and 45 brain slices were counted, respectively; n = 6 mice for each group. Results are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
To further verify the specifically cell-autonomous role of EphB2 and its signaling in adult rNSCs in SGZ and exclude the possible effects caused by hippocampus deficits during early development, we generated a Cre-dependent AAV with dominant-negative types of EphB2, AAV-DIO-EphB2-K661R-GFP, which blocks the catalytic function of EphB2. We injected these viruses into the DG area of Nestin-CreERT2 mice. To detect the proliferation of virus-infected rNSCs, the mice were traced by EdU and were perfused 1 week after tamoxifen injection (Fig. 5D). We found that the EphB2-K661R–infected rNSCs showed a higher level of proliferation than the control virus group (Fig. 5E). Furthermore, we traced the rNSCs and their progeny for 30 days and found that ~28% of GFP cells can be maintained in the rNSC stage in the AAV control group, while only ~16% of GFP+ rNSCs were left in the EphB2-K661R-GFP virus–injected DG area (Fig. 5, D and F). In the meantime, more DCX+ cells and neurons were observed in the EphB2-K661R-GFP virus–injected group (Fig. 5F). The data indicate that EphB2 kinase–dependent signaling is essential for quiescent NSC maintenance and newborn neuron generation in the hippocampus of adult mice.
To determine the roles of EphB2 kinase–dependent signals in rNSCs in NSC behavior during voluntary running trials, we injected AAV-DIO-EphB2-F620D-GFP, a constitutively active receptor form of EphB2 with ligand-independent catalytic activity (37), into the DG area of the Nestin-CreERT2 line (Fig. 5G). We found increased numbers of c-Fos+ cells in all groups after the 7-day running trial (Fig. 5H), while a fewer number of activated rNSCs in the mice injected with the EphB2-F620D virus were observed compared with the mice injected with the control virus (Fig. 5I). After the 30-day running trial, the lineage tracing experiments showed that the proportion of rNSCs and the neuronal progeny of rNSCs (DCX+ cells and neurons) significantly recovered in the EphB2-F620D virus–injected mice (Fig. 5J). The data suggest that EphB2 catalytic activity is sufficient to sustain the quiescence of adult rNSCs during voluntary running trials.
Neuronal ephrin-B3 and rNSC EphB2 work together to maintain rNSC quiescence
Last, to further test the requirement and sufficiency of the ephrin-B3–EphB2 signal in the regulation of quiescence of adult rNSC in vivo, we introduced EphB2−/−; Efnb3−/− double-knockout mice, which allowed us to examine the role of ligand in GCs and receptor in rNSCs specifically by rescuing neuronal ephrin-B3 or/and rNSC EphB2 signal in the neurogenic niche of adult hippocampus. We used AAV-CaMKII-ephrin-B3-mCherry to express ephrin-B3 specifically in dentate GCs and/or injected Cre-dependent AAV vectors (AAV-DIO-EphB2-GFP) into the DG region of Nestin-CreERT2 mice to express EphB2 in rNSCs (Fig. 6, A and B). We observed that the proportion of EdU-incorporated rNSCs, detected 7 days after labeling, and total rNSCs as well as the neuronal progeny of rNSCs (neurons), detected 30 days after lineage tracing, significantly recovered in the mice only if both neuronal ephrin-B3 and rNSC EphB2 were expressed simultaneously (Fig. 6, C to E). These results suggest that both ephrin-B3 in GCs and EphB2 in rNSCs are necessary and work together to sustain a quiescent NSC population in the DG region.
Fig. 6. Neuronal ephrin-B3 and rNSC EphB2 work together to maintain rNSC quiescence.
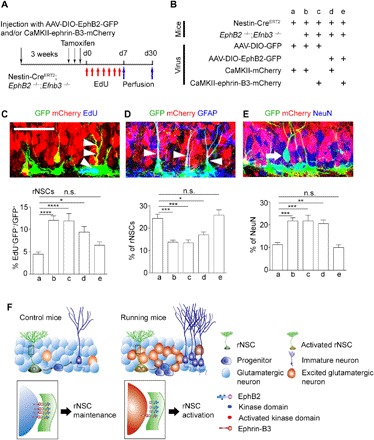
(A and B) Scheme depicting the experimental procedure of the specifically rescued expression of ephrin-B3 and EphB2 in excitatory neurons and rNSCs in adult EphB2−/−; Efnb3−/− double-knockout mice crossed with the Nestin-CreERT2 mouse line. (C to E) Confocal images showing virus-infected GFP+ rNSCs (arrowheads) in the DG colocalized with EdU (C), rNSCs (arrowheads) with GFAP (D), and neurons (arrow) with NeuN (E). Scale bar, 50 μm. Graphs show the proportion of EdU+ GFP+ rNSCs after EdU treatment (C), GFP+ rNSCs (D), and GFP+ NeuN (E) after 30-day lineage tracing in adult EphB2−/−; Efnb3−/− double-knockout mice crossed with the Nestin-CreERT2 mouse line. In groups a, b, c, d, and e, 3026, 3489, 2887, 3164, and 3165 GFP+ cells of 54, 54, 56, 57, and 56 brain slices were counted, respectively; n = 7 mice for each group. (F) Model for hippocampal quiescent NSCs activated by contacting excitatory neurons through direct neuron-rNSC interaction. In running mice, excited glutamatergic neurons directly interact with rNSCs, which leads to rNSC activation and thereby neurogenesis. This interaction is mediated by ephrin-B3–EphB2 signaling. Results are presented as means ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., not significant.
In summary, our study elucidates the physiological significance of GC excitability under environmental stimuli and indicates an inducible intercellular modulation via GC-rNSC contact. These results also indicate that the transcellular ephrin-B3–EphB2 signal pathway serves as a key-lock switch to control the functional shift of quiescent rNSCs responding to the activated GCs during voluntary running.
DISCUSSION
The continuous generation of new neurons in the DG is a prominent example of structural plasticity in the adult mammalian brain and is a process highly regulated by neural network activity. The GC hyperactivity can be elicited by seizures that activate rNSCs and gliogenesis, and this gives rise to rNSC depletion eventually but fails to generate newborn neurons (25, 26, 38). These results highlight not only the critical roles of GC overactivity in provoking a marked functional shift of adult rNSCs to glia but also its harmful impacts on hippocampal neurogenesis. In the present study, we provided positive evidence that physiological regulation of GC excitation by voluntary running profoundly affected quiescent rNSCs to promote generation of newborn neurons. We demonstrated that the regulation might be through a direct GC-rNSC interacting manner, which was different from the previous consensus in which secreted extracellular factors play a major role. The adult hippocampal rNSCs have a unique structure of long ramifying processes that extend from the SGZ toward the molecular layer. The 3D reconstruction of rNSCs and adjacent GC cells in adult DG, as shown in the present study, indicates that both the cell bodies and projections of rNSCs directly contact GC cells (Fig. 3A and movie S2). These structures were indispensable for rNSCs that the projections locating the molecular layer work as an antenna where they receive glutamatergic stimuli through structural and functional interactions with GCs in both developing and adult brains. Our study thus suggests the functional significance of the adjacent excitatory GCs in building the neurogenic niche for direct rNSC regulation (Fig. 6F).
The study also sheds light on the dual roles of the rNSC niche in NSC behaviors that involve either maintaining rNSCs in a quiescent state to prevent their depletion or directing their differentiation along the appropriate lineages (39, 40), and this is determined by a specific neuronal network responding selectively to physiological and pathological conditions (25, 26, 38). For instance, as a significant constituent part of the rNSC niche, dentate PV interneurons secrete GABA to maintain NSC quiescence (23, 24). Moreover, a recent study reveals that hippocampal mossy cells control aNSC quiescence through a dynamic balance between direct glutamatergic and indirect GABAergic pathways (16). In contrast to previous studies, we revealed, in the study, a dominant role of excitatory GC neurons in the niche that promoted adjacent rNSC activation and differentiation. These pieces of evidence together indicate that a balance of excitatory and inhibitory modulation is sophisticatedly orchestrated via coordination of various types of neurons in the niche that is crucial for the functional shift of rNSCs upon exposure to external environmental stimuli.
Our study further identified neuronal ephrin-B3 as an external negative regulator by analyzing previous sequencing results (28) and validating its protein expression in the excited DG area upon chemogenetic stimulation or voluntary running. We demonstrated that GC membrane–bound ephrin-B3 was the key protein that was down-regulated during long-time voluntary running, and this led to an attenuated EphB2 forward signaling into rNSCs, which is required for the activation of quiescent NSCs. These results are different from the ordinary roles played by Eph-ephrin signaling in the SVZ and SGZ (14, 15, 41), in which the effects of surrounding astrocyte/vascular cells, such as rNSC-niche cells, may not be adjustable in a timely manner and in which the cells respond directly to the environment stimuli. We thus provide a new mechanism of the ephrin-B3–EphB2 signal transduction that serves to couple external and internal environments, representing an environmental stimulus–dependent feedback from excitatory neurons to rNSCs in contrast to the feedback regulation proposed previously [for instance, the growth and differentiation factor 11–dependent negative feedback from neurons to neural progenitors in the olfactory epithelium (42) or epidermal growth factor–dependent feedback from neural progenitors to NSCs in the SVZ (18)]. In view of the colocalization of EphB2 in radial processes of rNSCs and ephrin-B3 in GCs (fig. S10B), the unique structure of rNSCs might be crucial for transducing the ephrin-B3–EphB2 signal pathway.
Mechanistically, the study indicates that EphB2-mediated forward signaling plays both essential and sufficient roles in maintaining the property of rNSCs through the kinase catalytic activity, whereas how the intercellular signaling is transduced within the rNSCs upon external stimuli remains an open question. In previous studies, mouse models based on PTEN deletion in the brain have provided information about the role of PTEN in cell proliferation and self-renewal of neural stem/progenitor cells (43), which has been performed either in embryonic NSCs (44) or in aNSCs from the SVZ of the lateral ventricles (43). Further evidence reveals that the Eph receptor physically interacts with PTEN to diminish its protein levels (45), while conversely, PTEN is also able to regulate stability and function of the EphB receptor (46). This suggests that PTEN-AKT signaling might be the downstream molecular pathway of the EphB receptor to regulate quiescent NSC behavior, which bridges an intracellular cell-autonomous pathway in adult rNSCs to transcellular stimuli from GCs in the neurogenic niche.
Together, our current results demonstrate that excitatory GC neurons may serve as a conveyor capable of relaying outside physiological stimuli to rNSCs through their membrane protein. This finding might lead to a number of implications in controlling adult hippocampal neurogenesis influenced by glutamatergic neurons and in developing optimal strategies depending on different levels of neuronal activity in treating brain disorders, such as Alzheimer’s disease (47) and schizophrenia (48), as they exhibit aberrant hippocampal neurogenesis. The study also indicates a direct neuron-rNSC interacting mechanism that is mediated by the transcellular ephrin-B3–EphB2 signal pathway, which would lead to a better understanding of how intercellular molecular signals were precisely transferred to regulate NSC behavior.
MATERIALS AND METHODS
Mice and sample preparation
EphB1−/− (49), EphB2−/− (35), EphB2LacZ (35), EphB2K661R (36), EphB2ΔVEV (36), EphB2F620D (37), Efnb3−/− (34), Nestin-GFP transgenic (27), CaMKII-Cre (50), Efnb3LacZ/lacZ (51), Td-tomato (Ai9) (52), Nestin-CreERT2 (53), and knockout and knock-in mice as well as genotyping methods have been described previously. These mutant mice were crossed into CD1 background. Animals were anesthetized with chloral hydrate (350 mg/kg) and perfused with 0.1 M phosphate-buffered saline (PBS) followed by 4% paraformaldehyde and 4% sucrose in PBS (pH 7.4). Brains were post-fixed overnight and sectioned at 30 μm using a vibratome. All experiments involving mice were carried out in accordance with the U.S. National Institutes of Health (NIH) Guide for the Care and Use of Animals under an Institutional Animal Care and Use Committee–approved protocol in a facility at the Shanghai Jiao Tong University School of Medicine that was approved by the Association for Assessment and Accreditation of Laboratory Animal Care. Parents and pups (10 to 11 pups per litter) were raised in animal facilities at a constant temperature (22°C) and on a 12-hour light/12-hour dark cycle. Access to food and water was unlimited. The day of birth was defined as postnatal day 0 (P0). All efforts were made to minimize the number of animals used and their suffering.
Voluntary running
Mice were randomly paired and assigned to two closed lines. In each subsequent generation, when the offspring of these pairs were 10 weeks old, they were housed individually with access to a running wheel for 7 and 30 days. For daily wheel running, mice were placed in a running wheel for 2 hours, which allowed them to voluntarily run. A control group of animals was maintained in a running wheel that does not rotate 2 hours a day for 7 days. A pulse of EdU was injected into each mouse after running both control and running mice. For the c-Fos counting, the mice were euthanized 1.5 hours after the last running trials. For other experiments, such as detecting rNSC proliferation and neurogenesis, the mice were euthanized 3 hours after the last EdU injection.
Fear conditioning
Mice were placed in a square chamber with a grid floor. On the first day (day 1), each mouse was habituated to the chamber for 3 min, and then an auditory cue was delivered (70 dB, 30 s) followed by a foot shock (0.85 mA, 2 s). Then, the mice were returned to their home cages. On the next day (day 2), the mice were exposed to the same chamber without any stimulus for 3 min. The contextual conditioning was assessed by recording freezing behavior during the 3-min exposure. After 2 hours, the mice were put in the same chamber for 3 min followed by a 30-s auditory cue. After 1.5 hours, we euthanized the mice to detect c-Fos expression.
Environmental enrichment
At the age of 10 weeks, a cohort of animals were housed in large cages [32 cm (width) by 47 cm (length) by 35 cm (height)] containing running wheels, colored cages, toys, and chewable materials 24 hours a day for 7 days. A control group of animals was maintained in standard housing conditions for 7 days. A pulse of EdU was injected into each mouse in the environmental enrichment group and the control group. After the 7-day treatment, we euthanized the mice to detect c-Fos expression.
Stereotaxic surgery
Young adult mice (6 weeks) were anesthetized with pentobarbital sodium (10 mg/kg). Virus was injected via a microsyringe (Hamilton, 1701 RN) and a microinjection pump (Hamilton) at a rate of 100 to 200 nl/min with the following coordinates: DG viral injection (0.5 μl): anteroposterior (AP), –2.1 mm; mediolateral (ML), ±1.7 mm; dorsoventral (DV), –2.2 mm. Mice were allowed to recover for 2 to 3 weeks from the surgery before in vivo experiments. For in vivo chemogenetics, CaMKII-Cre mice were injected with AAV2/9-CAG-DIO-hM3Dq/hM4Di-mCherry or AAV2/9-CAG-DIO-mCherry into the DG, and Nestin-CreERT2 mice were injected with AAV2/9-CaMKII-hM3Dq/hM4Di-mCherry or AAV2/9-CaMKII-mCherry into the DG. For in vivo lineage tracing, AAV2/9-CAG-DIO-GFP, AAV2/9-CAG-DIO-EphB2/EphB2-K661R-GFP, AAV2/9-CaMKII-mCherry, or AAV2/9-CaMKII-ephrin-B3-mCherry was injected into the DG of Nestin-CreERT2 mice.
Fiber photometry
Adult C57 mice were injected with AAV2/9-hSyn-GCaMP6s into the DG followed by optic fiber implantation 6 weeks postnatally. A 200-μm-diameter optical fiber was glued into a short cannula with the fiber tip extended approximately 2.5 mm out of the cannula. The preprocessed fiber was inserted through a small craniotomy made at the DG region (bregma coordinates: AP, –2.1 mm; ML, ±1.7 mm; DV, –2.05 mm). Last, the cannula was secured to the skull using dental cement. Mice were individually housed for at least 1 week to recover from fiber photometry. The rescue experiments were conducted 2 weeks after the virus was completely expressed.
For the running test, mice were first habituated to the fiber photometry apparatus for 30 min and then tested on a subsequent day. The fiber photometry was conducted by a fiber photometry system purchased from Thinker Tech Nanjing BioScience Inc. The analog voltage signals were digitized at 50 Hz. All the Ca2+ signals and behavior videos were synchronized offline with event marks. The Ca2+ signal data were segmented on the basis of behavior events with individual trials, which were marked at the time of experimental mice contact. We derived the values of fluorescence change (ΔF/F) by calculating (F − F0)/F0, where F0 is the baseline fluorescence signal averaged over a 2-s-long control time window. The ΔF/F data were presented with average plots with a shaded area indicating SEM.
Bioinformatics analysis
Gene Expression Omnibus (GEO) Series (GSE) 39697 dataset was downloaded from the GEO database of the National Center for Biotechnology Information. The raw RNA expression profile datasets were preprocessed using R 3.4.1 statistical software together with a Bioconductor package. In accordance with the R package, the robust multiarray average (RMA) algorithm was used to adjust for background intensities in the Affymetrix array data by including optical noise and nonspecific binding. The background-adjusted probe intensities were then converted into expression measures using the normalization and summarization methods encapsulated by the RMA algorithm. The k-nearest neighbor (KNN) algorithm was used to generate the missing values. We used the R package limma to assess the DEGs between 0 and 30 days of granular cell layer. DEGs were selected for which both the log2 fold change values were greater than 0.5 and P values were <0.05. We performed GO enrichment analysis with the DEGs using functional annotation tools in DAVID (Database for Annotation, Visualization and Integrated Discovery). The genes in three GO terms (membrane, GO: 0016020; receptor binding, GO: 0005102; and cell adhesion, GO: 0007155) were acquired from Mouse Genome Informatics of the Jackson laboratory. We compared the genes in the three GO terms and acquired 255 overlapping genes. Further, we compared the 255 genes with 1802 DEGs and finally acquired 46 overlapping genes. A Venn diagram was made using a Venn diagram tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Immunofluorescence and EdU staining
For immunofluorescence, coronal brain slices were blocked with permeable buffer (0.3% Triton X-100 in PBS) containing 10% donkey serum for an hour at room temperature and were incubated with primary antibodies in permeable buffer containing 2% donkey serum overnight at 4°C. The slices were then washed three times with PBS-T (0.1% Tween 20 in PBS) for 10 min each time and incubated with Alexa Fluor secondary antibodies (1:500, Molecular Probes) in the PBS buffer for 2 hours at room temperature. For primary antibodies, we used goat anti–β-gal (1:200, MP Biomedicals), chicken anti-GFP (1:3000; GFP-1020, Aves Labs), goat anti-SOX2 (1:200; sc-17320, Santa Cruz Biotechnology), goat anti-DCX (1:200; sc-8066, Santa Cruz Biotechnology), rabbit anti-NeuN (1:1000; 24307, Cell Signaling Technology), rabbit anti-DCX (1:200; 4604, Cell Signaling Technology), chicken anti-Tuj1 (1:1000; TUJ, Aves Labs), mouse anti-Nestin (1:200; 556309, BD Biosciences), goat anti-Ephb2 (1:1000; P54763, R&D Systems), goat anti-vGluT2 (1:500; ab101760, Abcam), goat anti-GAD56/67 (1:500; sc-7513, Santa Cruz Biotechnology), and rabbit anti-c-Fos (2250S, Cell Signaling Technology). For c-Fos immunostaining, mice were euthanized 1.5 hours after running, foot shock (0.8 mA, 2 s) and enriched environment exposure, or CNO administration. For EdU staining, the Click-iT Assay Kit with Alexa Fluor 647 (Life Technologies) was used according to the manufacturer’s instructions.
For quantifications of rNSCs and their progeny, coronal brain sections (30 μm) through the entire DG were collected in serial order. Immunostaining was performed on every sixth section encompassing the anterior to posterior end of the DG (from bregma −1.06 mm to −3.40 mm). Confocal images (Leica SP8) were used to analyze dual-immunofluorescence patterns. Quantitative analysis of labeled cells in the DG was performed with ImageJ software (RRID: SCR_003070). For each pair of primary antibodies, immune-reactive cells from representative sections containing the DG were counted and analyzed for dual labeling. Colocalization was determined by capturing confocal Z stack images and counting the number of neurons labeled by two markers in all focal planes.
For 3D reconstruction of confocal images, we captured confocal Z stack images every 0.28 μm consecutively in Nestin-GFP mice that had been injected with AAV-CaMKII-mCherry into the DG. This Z stack images were simulated to 3D images and movies using LAS X software (Leica Microsystems).
Western blotting
For Western blotting, the procedure was as previously performed (54), and the data were analyzed using NIH ImageJ software. For primary antibodies, we used mouse anti–glyceraldehyde phosphate dehydrogenase (1:3000; G8795, Sigma-Aldrich), goat anti-EphB2 (1:1000; P54763, R&D Systems), and rabbit anti-eB3 (1:1000; 34-3600, Invitrogen).
Quantitative real-time polymerase chain reaction
Total RNA was prepared from DG tissue or aNSCs of TRI Reagent (Sigma-Aldrich). RNAs were reverse-transcribed with High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems) according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (PCR) was performed using the SYBR Green Master Mix (Applied Biosystems) in a Fast 96-well System (Applied Biosystems). The relative copy number of β-actin RNA was quantified and used for normalization. We obtained the PCR primer sequences from the website https://pga.mgh.harvard.edu/primerbank/. Reverse transcription PCR primer sequences were as follows (5′→3′): Notch1_F: GATGGCCTCAATGGGTACAAG, Notch1_R: TCGTTGTTGTTGATGTCACAGT; Jagged1_F: ATGCAGAACGTGAATGGAGAG, Jagged1_R: GCGGGACTGATACTCCTTGAG; Itga3_F: CCTCTTCGGCTACTCGGTC, Itga3_R: CCAGTCCGGTTGGTATAGTCATC; Adam8_F: TGCTCAGCGTCTTATGGACAC, Adam8_R: AGGCCAAACCACTTCATACTG; Ptprf_F: TGCTCTCGTGATGCTTGGTTT, Ptprf_R: ATCCACGTAATTCGAGGCTTG; L1cam_F: AAAGGTGCAAGGGTGACATTC, L1cam_R: TCCCCACGTTCCTGTA-GGT; VCAM1_F: TTGGGAGCCTCAACGGTACT, VCAM1_R: GCAATCGTTTTGTATTCAGGGGA; Lcam2_F: CAGTTCCGAGGTCTACACT T, Lcam2_R: TGAATCGGGAGTCTTCCGAAAA; Flot1_F: CGGAGGCCGAGTGTTTGTC, Flot1_R: CCGTGGCGGGTATAAACCTTT; N-Cad_F: AGGCTTCTGGTGAAATTGCAT, N-Cad_R: GTCCACCTTGAAATCTGCTGG; Pvr_F: GGGTGGGGATATACGTGTGC, Pvr_R: GAGATGCGTTCCTCAGATCCT; Epha4_F: TGGAATTTGCGACGCTGTCA, Epha4_R: CACTTCCTCCCACCCTCCTT; Efnb1_F: TGTGGCTATGGTCGTGCTG, Efnb1_R: TCTTCGGGTAGATCACCAAGC; Efnb3_F: GAGGGCGGTTACGT-GCTTTAT, Efnb3_R: GGGCTGTATTCCTGGAACTTGAT; EphB1_F: CCTCCTCCTATGGACTGCCC, EphB1_R: AAGGCCGTGAAGTCTGGGATA; EphB2_F: GCCGTGGAAGAAACCCTGAT, EphB2_R: GTTCATGTTCTCGTCGTAGCC; EphB3_F: CATGAATCCTATCCGCACGTATC, EphB3_R: GCTGTTACAGTCTCTTACGGTGA; β-actin_F: GCTCTTTTCCAGCCTTCCTT, β-actin_R: TGATCCACATCTGCTGGAAG.
Statistical analysis
The results are presented as means ± SEM. Statistical differences were determined by Student’s t test for two-group comparisons or analysis of variance (ANOVA) followed by Tukey test for multiple comparisons with more than two groups.
Supplementary Material
Acknowledgments
We thank H. Zeng for providing CAGG-tdTomato transgenic mice, J. Gao and Y. Li for providing CaMKII-Cre transgenic mice, M. Henkemeyer for all EphB and Efnb3 mutant mice lines, Y. Xie and Z. Yang for Nestin-CreERT2 mice, and G.-N. Xu and S. Chen for laboratory technique support. Funding: This research was supported by the National Basic Research Program of China (973 Program, 2014CB965002) to N.-J.X., the National Natural and Science Foundation of China (31371097, 81870820, and 31671062 to N.-J.X. and 81471151 to S.S.), Grants of Shanghai Brain-Intelligence Project from the Science and Technology Commission of Shanghai Municipality (STCSM) (16JC1420500), the STCSM (18JC1420302), and the Recruitment Program of Global Experts for Young Professionals (to N.-J.X.). Author contributions: J.D., L.-N.H., and X.-D.L. performed morphological, histological, and animal experiments. Y.-B.P. performed bioinformatics analysis. X.-R.W. performed fiber photometry experiments. J.D., D.-F.F., T.-L.X., S.S., and N.-J.X. designed the experiments. J.D., S.S., and N.-J.X. analyzed the data and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaav4416/DC1
Fig. S1. Activated GC neurons after treatment of contextual fear, enriched environment exposure, or voluntary running, respectively.
Fig. S2. Unchanged total GFP+ NSCs and increased Ki67+ rNSCs and ANPs after running.
Fig. S3. Increased DCX+ cells and Tbr2+ cells in the hippocampus after voluntary running.
Fig. S4. Excited DG glutamatergic neurons but not GABAergic neurons in voluntary running.
Fig. S5. AAV-infected vGluT2+-specific DG neurons.
Fig. S6. c-Fos+ neurons in the DG region after CNO treatment in various doses.
Fig. S7. Effects of CNO control with various doses.
Fig. S8. Analysis of sequencing results.
Fig. S9. Ephrin-B3 ligand prevents proliferation/differentiation of aNSCs and their transition to neurons.
Fig. S10. EphB2 kinase–dependent signaling is required for the maintenance of quiescent rNSCs.
Movie S1. Voluntary running behavior of a mouse in the running wheel.
Movie S2. In vivo fiber photometry of Ca2+ signal of DG granule neurons during running trials.
Movie S3. 3D reconstruction of confocal images of rNSCs and GCs.
REFERENCES AND NOTES
- 1.Eriksson P. S., Perfilieva E., Björk-Eriksson T., Alborn A.-M., Nordborg C., Peterson D. A., Gage F. H., Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Ming G.-l., Song H., Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 70, 687–702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuentealba L. C., Obernier K., Alvarez-Buylla A., Adult neural stem cells bridge their niche. Cell Stem Cell 10, 698–708 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda S., Kato F., Tozuka Y., Yamaguchi M., Miyamoto Y., Hisatsune T., Two distinct subpopulations of nestin-positive cells in adult mouse dentate gyrus. J. Neurosci. 23, 9357–9366 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suh H., Consiglio A., Ray J., Sawai T., D'Amour K. A., Gage F. H., In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell 1, 515–528 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonaguidi M. A., Wheeler M. A., Shapiro J. S., Stadel R. P., Sun G. J., Ming G.-l., Song H., In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akers K. G., Martinez-Canabal A., Restivo L., Yiu A. P., De Cristofaro A., Hsiang H.-L., Wheeler A. L., Guskjolen A., Niibori Y., Shoji H., Ohira K., Richards B. A., Miyakawa T., Josselyn S. A., Frankland P. W., Hippocampal neurogenesis regulates forgetting during adulthood and infancy. Science 344, 598–602 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Deng W., Aimone J. B., Gage F. H., New neurons and new memories: How does adult hippocampal neurogenesis affect learning and memory? Nat. Rev. Neurosci. 11, 339–350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder J. S., Soumier A., Brewer M., Pickel J., Cameron H. A., Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature 476, 458–461 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clelland C. D., Choi M., Romberg C., Clemenson G. D. Jr., Fragniere A., Tyers P., Jessberger S., Saksida L. M., Barker R. A., Gage F. H., Bussey T. J., A functional role for adult hippocampal neurogenesis in spatial pattern separation. Science 325, 210–213 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christian K. M., Song H., Ming G.-l., Functions and dysfunctions of adult hippocampal neurogenesis. Annu. Rev. Neurosci. 37, 243–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mira H., Andreu Z., Suh H., Lie D. C., Jessberger S., Consiglio A., San Emeterio J., Hortigüela R., Marqués-Torrejón M. Á., Nakashima K., Colak D., Götz M., Fariñas I., Gage F. H., Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Jang M.-H., Bonaguidi M. A., Kitabatake Y., Sun J., Song J., Kang E., Jun H., Zhong C., Su Y., Guo J. U., Wang M. X., Sailor K. A., Kim J.-Y., Gao Y., Christian K. M., Ming G.-l., Song H., Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashton R. S., Conway A., Pangarkar C., Bergen J., Lim K.-I., Shah P., Bissell M., Schaffer D. V., Astrocytes regulate adult hippocampal neurogenesis through ephrin-B signaling. Nat. Neurosci. 15, 1399–1406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ottone C., Krusche B., Whitby A., Clements M., Quadrato G., Pitulescu M. E., Adams R. H., Parrinello S., Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat. Cell Biol. 16, 1045–1056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh C.-Y., Asrican B., Moss J., Quintanilla L. J., He T., Mao X., Cassé F., Gebara E., Bao H., Lu W., Toni N., Song J., Mossy cells control adult neural stem cell quiescence and maintenance through a dynamic balance between direct and indirect pathways. Neuron 99, 493–510.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama R., Ohtsuka T., Shimojo H., Imayoshi I., Dynamic Notch signaling in neural progenitor cells and a revised view of lateral inhibition. Nat. Neurosci. 11, 1247–1251 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Aguirre A., Rubio M. E., Gallo V., Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467, 323–327 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nilsson M., Perfilieva E., Johansson U., Orwar O., Eriksson P. S., Enriched environment increases neurogenesis in the adult rat dentate gyrus and improves spatial memory. J. Neurobiol. 39, 569–578 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Kempermann G., Kuhn H. G., Gage F. H., More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Kirby E. D., Friedman A. R., Covarrubias D., Ying C., Sun W. G., Goosens K. A., Sapolsky R. M., Kaufer D., Basolateral amygdala regulation of adult hippocampal neurogenesis and fear-related activation of newborn neurons. Mol. Psych. 17, 527–536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Praag H., Kempermann G., Gage F. H., Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2, 266–270 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Song J., Zhong C., Bonaguidi M. A., Sun G. J., Hsu D., Gu Y., Meletis K., Huang Z. J., Ge S., Enikolopov G., Deisseroth K., Luscher B., Christian K. M., Ming G.-l., Song H., Neuronal circuitry mechanism regulating adult quiescent neural stem-cell fate decision. Nature 489, 150–154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bao H., Asrican B., Li W., Gu B., Wen Z., Lim S.-A., Haniff I., Ramakrishnan C., Deisseroth K., Philpot B., Song J., Long-range GABAergic inputs regulate neural stem cell quiescence and control adult hippocampal neurogenesis. Cell Stem Cell 21, 604–617.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hüttmann K., Sadgrove M., Wallraff A., Hinterkeuser S., Kirchhoff F., Steinhäuser C., Gray W. P., Seizures preferentially stimulate proliferation of radial glia-like astrocytes in the adult dentate gyrus: Functional and immunocytochemical analysis. Eur. J. Neurosci. 18, 2769–2778 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Sierra A., Martín-Suárez S., Valcárcel-Martin R., Pascual-Brazo J., Aelvoet S. A., Abiega O., Deudero J. J., Brewster A. L., Bernales I., Anderson A. E., Baekelandt V., Maletić-Savatić M., Encinas J. M., Neuronal hyperactivity accelerates depletion of neural stem cells and impairs hippocampal neurogenesis. Cell Stem Cell 16, 488–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu T.-S., Dandekar M., Monteggia L. M., Parada L. F., Kernie S. G., Temporally regulated expression of Cre recombinase in neural stem cells. Genesis 41, 147–153 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Miller J. A., Nathanson J., Franjic D., Shim S., Dalley R. A., Shapouri S., Smith K. A., Sunkin S. M., Bernard A., Bennett J. L., Lee C.-K., Hawrylycz M. J., Jones A. R., Amaral D. G., Sestan N., Gage F. H., Lein E. S., Conserved molecular signatures of neurogenesis in the hippocampal subgranular zone of rodents and primates. Development 140, 4633–4644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giagtzoglou N., Ly C. V., Bellen H. J., Cell adhesion, the backbone of the synapse: “Vertebrate” and “invertebrate” perspectives. Cold Spring Harb. Perspect. Biol. 1, a003079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grunwald I. C., Korte M., Wolfer D., Wilkinson G. A., Unsicker K., Lipp H.-P., Bonhoeffer T., Klein R., Kinase-independent requirement of EphB2 receptors in hippocampal synaptic plasticity. Neuron 32, 1027–1040 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Xu N.-J., Henkemeyer M., Ephrin-B3 reverse signaling through Grb4 and cytoskeletal regulators mediates axon pruning. Nat. Neurosci. 12, 268–276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knöll B., Drescher U., Ephrin-As as receptors in topographic projections. Trends Neurosci. 25, 145–149 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Pasquale E. B., Eph-ephrin bidirectional signaling in physiology and disease. Cell 133, 38–52 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Xu N.-J., Sun S., Gibson J. R., Henkemeyer M., A dual shaping mechanism for postsynaptic ephrin-B3 as a receptor that sculpts dendrites and synapses. Nat. Neurosci. 14, 1421–1429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkemeyer M., Orioli D., Henderson J. T., Saxton T. M., Roder J., Pawson T., Klein R., Nuk controls pathfinding of commissural axons in the mammalian central nervous system. Cell 86, 35–46 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Genander M., Halford M. M., Xu N. J., Eriksson M., Yu Z., Qiu Z., Martling A., Greicius G., Chumley M. J., Zdunek S., Wang C., Holm T., Goff S. P., Pettersson S., Pestell R. G., Henkemeyer M., Frisén J., Dissociation of EphB2 signaling pathways mediating progenitor cell proliferation and tumor suppression. Cell 139, 679–692 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmberg J., Genander M., Halford M. M., Anneren C., Sondell M., Chumley M. J., Silvany R. E., Henkemeyer M., Frisén J., EphB receptors coordinate migration and proliferation in the intestinal stem cell niche. Cell 125, 1151–1163 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Lugert S., Basak O., Knuckles P., Häaussler U., Fabel K., Götz M., Haas C. A., Kempermann G., Taylor V., Giachino C., Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6, 445–456 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Fuchs E., Tumbar T., Guasch G., Socializing with the neighbors: Stem cells and their niche. Cell 116, 769–778 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Miller F. D., Gauthier-Fisher A., Home at last: Neural stem cell niches defined. Cell Stem Cell 4, 507–510 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Conover J. C., Doetsch F., Garcia-Verdugo J.-M., Gale N. W., Yancopoulos G. D., Alvarez-Buylla A., Disruption of Eph/ephrin signaling affects migration and proliferation in the adult subventricular zone. Nat. Neurosci. 3, 1091–1097 (2000). [DOI] [PubMed] [Google Scholar]
- 42.Wu H.-H., Ivkovic S., Murray R. C., Jaramillo S., Lyons K. M., Johnson J. E., Calof A. L., Autoregulation of neurogenesis by GDF11. Neuron 37, 197–207 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Gregorian C., Nakashima J., Le Belle J., Ohab J., Kim R., Liu A., Smith K. B., Groszer M., Garcia A. D., Sofroniew M. V., Carmichael S. T., Kornblum H. I., Liu X., Wu H., Pten deletion in adult neural stem/progenitor cells enhances constitutive neurogenesis. J. Neurosci. 29, 1874–1886 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groszer M., Erickson R., Scripture-Adams D. D., Lesche R., Trumpp A., Zack J. A., Kornblum H. I., Liu X., Wu H., Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science 294, 2186–2189 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Brisbin S., Liu J., Boudreau J., Peng J., Evangelista M., Chin-Sang I., A role for C. elegans Eph RTK signaling in PTEN regulation. Dev. Cell 17, 459–469 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez S., Huynh-Do U., Phosphatase and tensin homolog regulates stability and activity of EphB1 receptor. FASEB J. 27, 632–644 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Mu Y., Gage F. H., Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol. Neurodegener. 6, 85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J. Y., Liu C. Y., Zhang F., Duan X., Wen Z., Song J., Feighery E., Lu B., Rujescu D., St. Clair D., Christian K., Callicott J. H., Weinberger D. R., Song H., Ming G.-l., Interplay between DISC1 and GABA signaling regulates neurogenesis in mice and risk for schizophrenia. Cell 148, 1051–1064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams S. E., Mann F., Erskine L., Sakurai T., Wei S., Rossi D. J., Gale N. W., Holt C. E., Mason C. A., Henkemeyer M., Ephrin-B2 and EphB1 mediate retinal axon divergence at the optic chiasm. Neuron 39, 919–935 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Tsien J. Z., Chen D. F., Gerber D., Tom C., Mercer E. H., Anderson D. J., Mayford M., Kandel E. R., Tonegawa S., Subregion- and cell type–restricted gene knockout in mouse brain. Cell 87, 1317–1326 (1996). [DOI] [PubMed] [Google Scholar]
- 51.Yokoyama N., Romero M. I., Cowan C. A., Galvan P., Helmbacher F., Charnay P., Parada L. F., Henkemeyer M., Forward signaling mediated by ephrin-B3 prevents contralateral corticospinal axons from recrossing the spinal cord midline. Neuron 29, 85–97 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R., Lein E. S., Zeng H., A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagace D. C., Whitman M. C., Noonan M. A., Ables J. L., DeCarolis N. A., Arguello A. A., Donovan M. H., Fischer S. J., Farnbauch L. A., Beech R. D., DiLeone R. J., Greer C. A., Mandyam C. D., Eisch A. J., Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27, 12623–12629 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun S., Zhang H., Liu J., Popugaeva E., Xu N.-J., Feske S., White C. L. III, Bezprozvanny I., Reduced synaptic STIM2 expression and impaired store-operated calcium entry cause destabilization of mature spines in mutant presenilin mice. Neuron 82, 79–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/2/eaav4416/DC1
Fig. S1. Activated GC neurons after treatment of contextual fear, enriched environment exposure, or voluntary running, respectively.
Fig. S2. Unchanged total GFP+ NSCs and increased Ki67+ rNSCs and ANPs after running.
Fig. S3. Increased DCX+ cells and Tbr2+ cells in the hippocampus after voluntary running.
Fig. S4. Excited DG glutamatergic neurons but not GABAergic neurons in voluntary running.
Fig. S5. AAV-infected vGluT2+-specific DG neurons.
Fig. S6. c-Fos+ neurons in the DG region after CNO treatment in various doses.
Fig. S7. Effects of CNO control with various doses.
Fig. S8. Analysis of sequencing results.
Fig. S9. Ephrin-B3 ligand prevents proliferation/differentiation of aNSCs and their transition to neurons.
Fig. S10. EphB2 kinase–dependent signaling is required for the maintenance of quiescent rNSCs.
Movie S1. Voluntary running behavior of a mouse in the running wheel.
Movie S2. In vivo fiber photometry of Ca2+ signal of DG granule neurons during running trials.
Movie S3. 3D reconstruction of confocal images of rNSCs and GCs.


