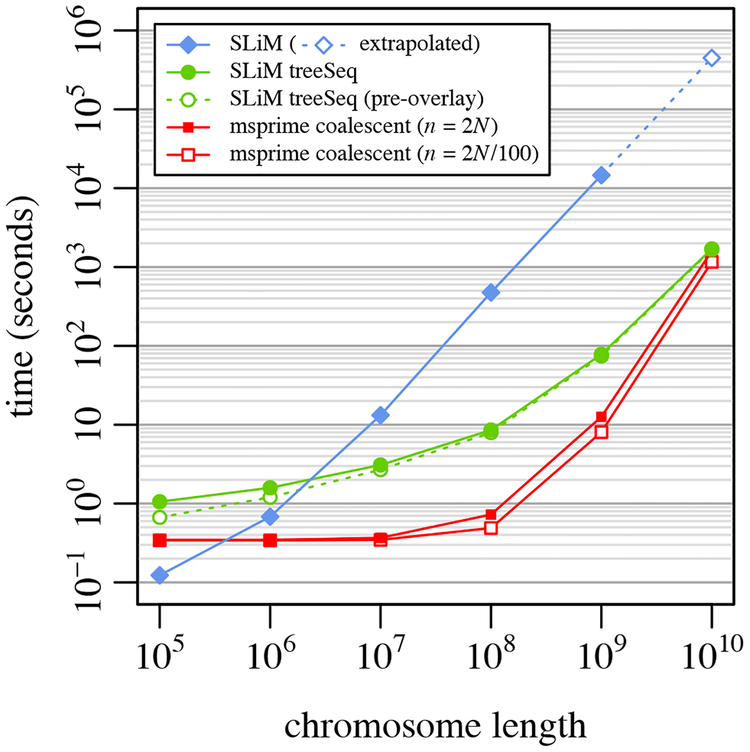
Figure 2.
A speed comparison between SLiM without tree-sequence recording, SLiM with tree-sequence recording and mutation overlay, and msprime’s coalescent simulation for a simple neutral model (Example 1; see text for model description). Each point represents the mean runtime across 10 replicates using different random number seeds; bars showing standard error of the mean would be smaller than the size of the plotted points in all cases. Runs for SLiM without tree-sequence recording (filled blue diamonds) were not conducted for L = 1010 because the memory usage was prohibitive, so a linear extrapolation is shown (hollow blue diamond). Runs for SLiM with tree-sequence recording and mutation overlay (filled green circles) are subdivided here to show the runtime for SLiM alone, prior to mutation overlay (hollow green circles), illustrating that the time for mutation overlay is negligible. The runtimes for the msprime coalescent for a full population sample of n = 2N = 1000 (filled red squares) and for a sample of size n = 2N/100 = 10 (hollow red squares) are both shown. Note that the x and y axes are both on a log scale.