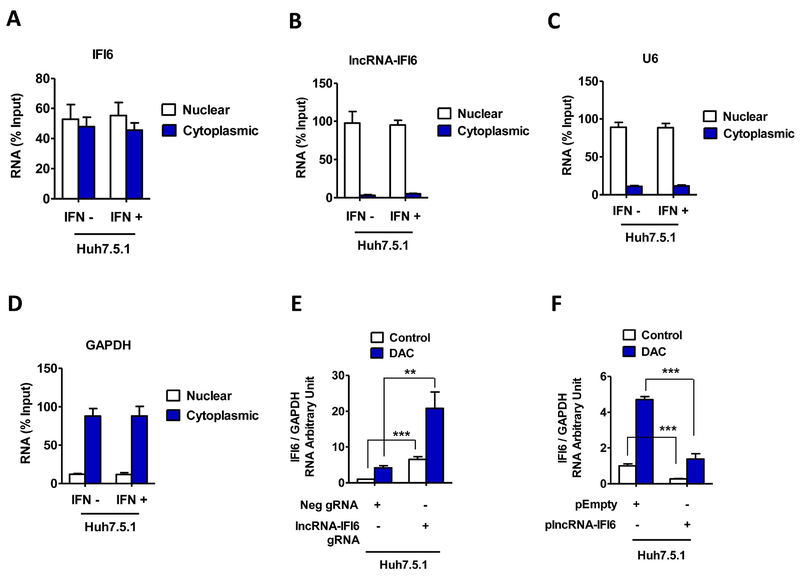
Figure 4. lncRNA-IFI6 is a nuclear transcript.
Huh7.5.1 cells were treated with IFNα (30 IU/ mL) for 24 hrs. Cytoplasmic and nuclear fractions of the cells were separated. The RNA levels of lncRNA-IFI6 and IFI6 were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. The total RNA was used as input control. Data are shown as percent (%) input. lncRNA-IFI6 gRNA, Neg gRNA, plncRNA-IFI6 and pEmpty stably overexpressing cells were treated with or without 10 μM 5-Aza-2’-deoxycytidineDNA (DAC) (methyltransferase inhibitor decitabine) for 48 hrs. Total RNA was isolated. IFI6 mRNA was assessed by qRT-PCR. (A) IFI6 mRNA was detected in both nuclear and cytoplasmic fractions in Huh7.5.1 cells. (B) lncRNA-IFI6 is a nuclear transcript. The majority of lncRNA-IFI6 was found in the nuclear fraction of Huh7.5.1 cells. (C) RNA levels of U6 (nuclear marker) were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. Total RNA was used as input control. Data are shown as percentage input. (D) RNA levels of GAPDH (cytoplasmic marker) were assessed by qRT-PCR in nuclear and cytoplasmic fractions from Huh7.5.1 cells. (E) lncRNA IFI6 gRNA or DNA methyltransferase inhibitor decitabine (DAC) (15 μM) treatment increased IFI6 mRNA levels in Huh7.5.1 cells. (F) Overexpression of plncRNA IFI6 blocked DAC treatment-induced IFI6 mRNA enhancement in Huh7.5.1 with or without IFNα, respectively.