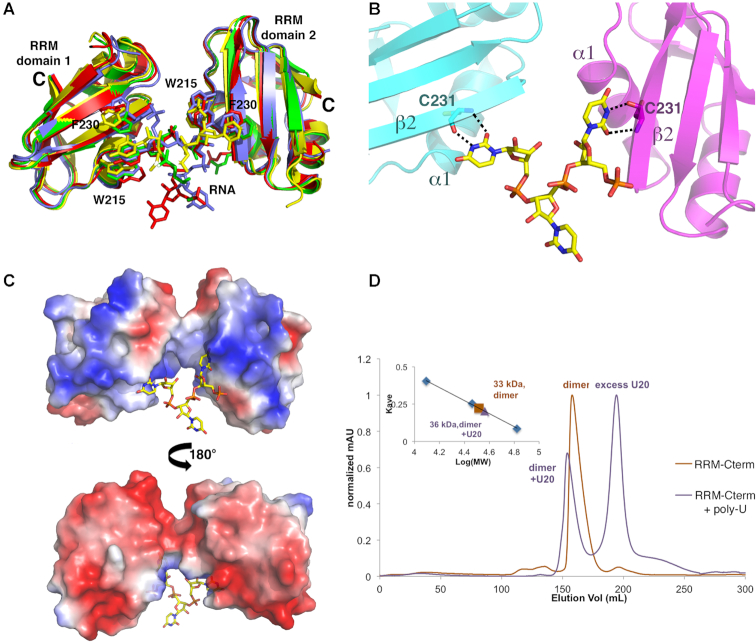
Figure 3.
Mechanism of uridine RNA recognition by the TbRGG2 RRM domain. (A) Overlay of the distinct RRM-RNA complexes in the crystallographic ASU showing the same overall binding mode involving an RRM dimer. (B) Close up of uridine recognition by the TbRGG2 RRM domains. Each uridine N3 and O2 atom is read by the backbone carbonyl and amide nitrogen of Cys231, respectively. (C) Electrostatic surface representation of the two RRMs binding to U4 where red and blue represent electronegative and electropositive regions, respectively. (D) SEC experiments analyzing the RRM-Cterm with and without U20 RNA. The RRM-Cterm alone (orange chromatogram) runs at a molecular weight (MW) of 33 kDa and when U20 is present, the largest peak (purple) corresponds to a MW of 36 kDa, which is consistent with an RRM-Cterm dimer bound to one strand of RNA (RRM-Cterm dimer MW = 31 kDa and U20 MW = 6 kDa). The inset is the curve used to determine MW, where the y-axis is elution volume normalized for the column volume and the x-axis is log(MW).