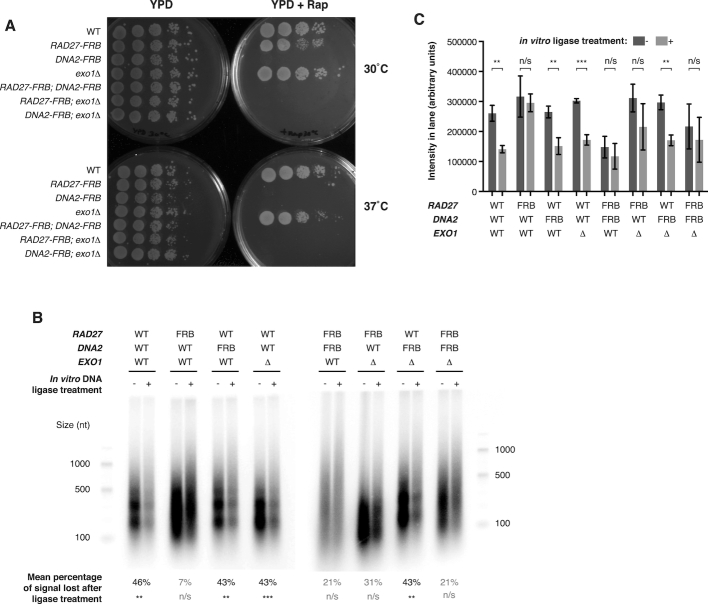
Figure 1.
Enrichment of ligation-competent Okazaki fragments from strains lacking both lagging-strand nucleases and DNA ligase I. (A) Spot tests of indicated strains on YPD ± 2 μg/ml rapamycin at 30 or 37°C, as indicated, for single and double mutant strains analyzed in this study. Addition of rapamycin depletes FRB-tagged proteins from the nucleus. (B) End-labeled Okazaki fragments obtained after 1 h rapamycin treatment to deplete DNA ligase I and FRB-tagged nucleases. Where indicated, DNA was treated with T4 DNA ligase after purification but before labeling to assess the proportion of Okazaki fragments poised for ligation. WT denotes an otherwise wild-type CDC9-FRB strain. The percentage of signal lost upon ligase treatment, calculated from three replicates and shown graphically in (C), is indicated under each pair of lanes. In both (B) and (C), P values for differences between paired samples with and without ligase were calculated by t-test. *** denotes P < 0.001, ** denotes P < 0.01, n/s denotes P > 0.05. (C) Quantification of Okazaki fragment end-labeling signal from (B). The cumulative signal from the region corresponding to sizes <1000 nt was calculated for three replicate experiments, of which (B) is a representative example. Mean ± SD is plotted and P values are as in (B).