Figure 7.
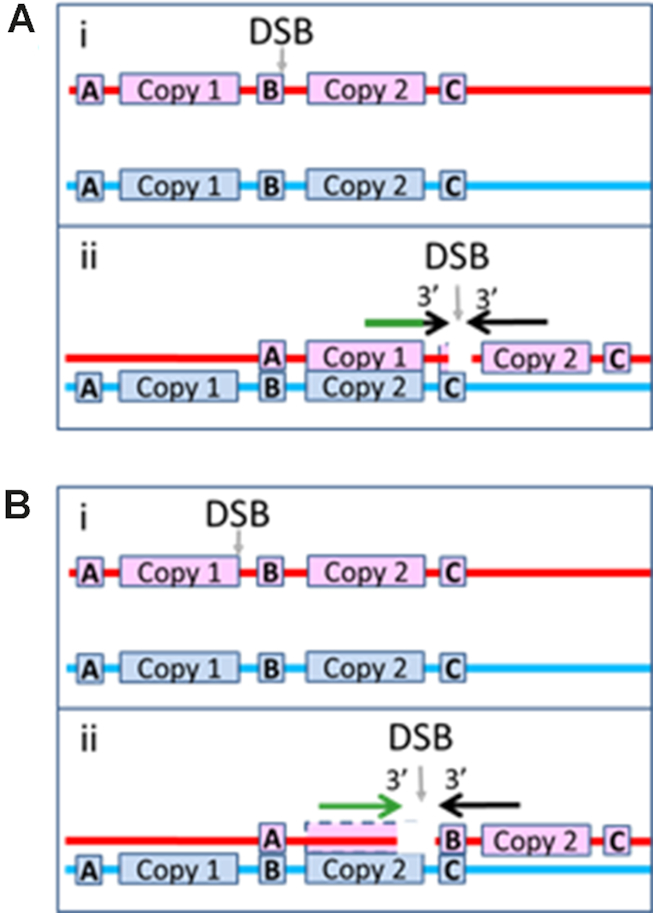
Illustration of how genomic rearrangement can be reduced for DSBs that occur outside of repeats. Same as Figure 1B, but the DSB occurs outside of a repeat. (A) The right filament is heterologous to the sequence to the right of Copy 2, so it cannot form a heteroduplex product by pairing with the dsDNA to the right of Copy 2. The left filament includes enough of Copy 1 to form a very stable N bp heteroduplex product when paired with the corresponding bases in Copy 2, but the black region near the tip of the arrow shows that the N homologous bp are separated from the 3′ of the filament by M3′ bases that are not homologous to the corresponding bases in Copy 2. Those M3′ mismatched bases could inhibit the progress of strand exchange beyond the sequence matched N bp heteroduplex because strand exchange proceeds less effectively through regions that contain mismatches. (B) i. A DSB occurs just before the right edge of Copy 1. ii. The left filament can form a long sequence matched heteroduplex product at the 3′ end of the initiating strand, but the right filament cannot form a stable heteroduplex in the region to the right of Copy 2. If synthesis triggered by one filament can produce an irreversible strand exchange product, then this geometry can produce genomic rearrangement; however, if both filaments must trigger synthesis that completes two dsDNAs, this interaction would not produce genomic rearrangement, even though the right filament could trigger synthesis.
