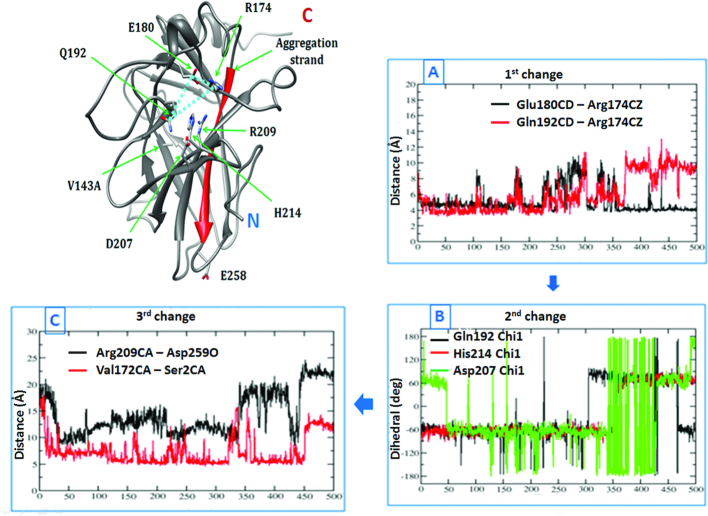
Figure 8.
A change in the interaction pattern of a subset of residues across MD simulations causes ‘opening’ of the S6–S7 strand in the destabilizing mutants. Structure of the V143A DBD is shown on the left. Plots (A), (B) and (C) on the right show how a perturbation in the reaction coordinate (involving the residues labelled in the structure on the left) (dihedral/distance) trigger a change in another reaction coordinate across a 500 ns MD (x-axis). Plot (A) shows that the triad of three interacting residues (R174, E180 and Q192) is destabilized. Plot (B) records the following changes in the side chain dihedrals of the vicinal residues (Q192, D207, H214). Subsequently the S6–S7 strand assumes an ‘open’ conformation (Plot (C)) displacing the N-terminus into a non-native conformation (as shown in the structure on the left).