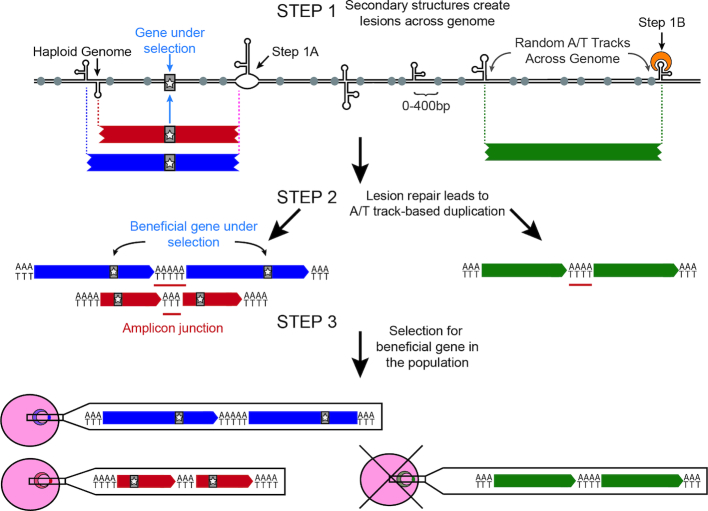
Figure 5.
Model of CNV development and selection in Plasmodium falciparum. In Step 1, DNA hairpins trigger double strand breaks throughout the P. falciparum genome presumably by either halting replication fork progression (Step 1A) or recognition by hairpin-binding proteins (Step 1B). In Step 2, long A/T tracks (gray circles) within 400 bp of the double strand break are utilized as microhomology for error-prone repair pathways to generate CNVs (blue, red and green bars). CNV breakpoints (vertical dotted lines) are generated semi-randomly across the genome but more stable hairpins are more likely to generate recurrent breakpoints (purple dotted line). De novo CNVs can either contain beneficial genes (gray bar with star) or those unrelated to the selection. New CNVs are generated frequently and could randomly occur throughout the highly repetitive P. falciparum genome (green bar), but may increase under selective pressure (see Discussion). In Step 3, selection (i.e. drug or fitness effects) enriches for beneficial CNVs (blue and red parasites) and purges deleterious CNVs (green parasite) from the population.